Using RFLP-PCR, Mini Sequencing and STR Techniques in Preimplantation Diagnosis of Spinal Muscular Dystrophy in Vietnam
Introduction
Spinal Muscular Atrophy (SMA) is one of the most common autosomal recessive disorders in children after Duchenne muscular dystrophy. SMA is characterized by degeneration and loss of motor neurons of the spinal cords anterior horn, hallmark symptoms in proximal symmetrical muscle weakness, and atrophy. A multipleethnic study reported that the overall SMA carrier prevalence was 1 in 54, and this disease is affecting approximately 1 in 11,000 live births [1,2]. SMA patients can be classified into three types according to the motor dysfunction and age of onset category: type 1 is the most severe group with the early onset before six months, the children often cannot be alive to 2 years old; type 2 is a moderate form, with onset before 18 months and diminished life expectancy; type 3 is the mildest form with later onset and better life expectancy [3]. Subsequent modifications referred to a type 4 adjunct for adult-onset patients and a type 0 for prenatal onset making deaths at neonatal [4,5]. There is no effective therapy for SMA however broadening our knowledge about the molecular genetics of the disease is resultant to develop potential treatment strategy as well as Preimplantation Genetic Tests Monogenic [6- 10]. Estimated, 95% of SMA patients are caused by homozygous deletions of exon 7 in the SMN1 gene (Survival Motor Neuron 1) or SMNt (the telomeric copy). The gene is presented in a complex region of 5q13 which also contains SMN2 (survival motor neuron 2) or SMNc (the centromeric copy), a homologous pseudogene of SMNt [11-14].
These couple genes differ in five intronic and three exonic nucleotides, one of which is encoded in exon 7; they allow SMNt to be separated from SMN2 and are currently being used as a genetic diagnostic tool for SMA [15]. Preimplantation Genetic Tests Monogenic (PGT-M) is now more popular and plays a crucial role in alternative to prenatal diagnosis for couples with a background of recessive genetic disorders. For spinal muscular atrophy, PGT-M has previously been applied in several centers. The procedure requires obtaining DNA samples from biopsied single cells at the cleavage stage, followed by genetic analysis for selecting an unaffected embryo to transfer into intrauterine. The principle of the analyzing process was detecting homozygous deletion of exon 7 of the SMNt gene by either nested PCR followed by restriction enzyme digestion or by fluorescent PCR with allele-specific amplification [16,17]. Up to date, a PGT-M protocol of SMA has various changes based on advent molecular developments; therefore, in this study, a PGT-M protocol was established with some modifications focused on blastocyst biopsy, DNA amplification for the SMNt gene mutation [18,19].
Materials and Methods
Lymphocytes Preparation
Human lymphocytes from 17 SMA patients with the homozygous exon 7 deletions of the SMNt gene and their heterozygous carrier parents were isolated from fresh blood. A total of 400 μl of fresh peripheral blood was taken and diluted 1:1 with sterile Phosphate- Buffered Saline (PBS) then mixed gently with a pipette. Afterward, 600 μl of Ficoll-Paque PREMIUM (Cytiva, USA, LOT No.10265111, EXP. Date: 01/2022) was overlaid with the diluted blood sample and centrifuged following the manufacturer’s instructions. The aspirated lymphocyte layer was washed through three changes of sterile PBS before being resuspended. Then, a stereoscopic microscope was used to collect lymphocytes by diluting them through droplets of PBS. These single cells were picked up, washed through another two drops of PBS buffer, and put into PCR tubes containing 2 μl PBS.
Blastocyst Embryos Biopsy
Twenty-nine high score blastocyst embryos remaining after completing IVF treatments of different couples who did not have SMA medical history were prepared. They were selected and transferred from liquid nitrogen storage container to thawing solution maintaining at 37°C, then placed in a Global culture medium droplet (Global, Life Global, USA) in a culture dish. All the biopsy process was performed on the heated stage of an inverted microscope equipping with micromanipulation tools. Each embryo was oriented on the holding pipette to visualize the Inner Cell Mass (ICM) at the position of 7 o’clock. A diode laser applied a trophectoderm biopsy with opening Zona Pellucida (ZP) at the blastocyst stage. A 8 μm hole in the zona pellucida was made by a cumulative laser pulse from the outer inwards to avoid damaging the ICM. Then, the biopsy pipette was pressed against the zona carefully to detach the TE cells from the internal surface of ZP. Several 5 – 10 cells were aspirated into the pipette and washed with PBS 1X and 1% PVP solution, then contained in the 0.2 mL PCR tube and stored at −20°C.
Whole-Genome Amplification
The DNA from both lymphocytes and biopsied blastomeres were amplified with REPLI-g® Single Cell Kit (QIAGEN, Germany, LOT No.169023130, EXP. Date: 05/2022) and diluted with nuclease-free water to reach a concentration of around 20 ng/μL. The concentration and purification were calculated with a Spectra Max Quick Drop. Therefore, the amplified DNA collected from the embryonic cells was qualified to be used in the research. DNA samples were stored at −20°C.
Double Methods for Detecting the Mutation
Restriction Fragment Length Polymorphism (RFLP): The products from whole genome amplification were used as templates to amplify the exon 7 of the SMNt gene with polymerase chain reaction (PCR). The primers used for the reaction were previously described in 2001 with some modifications: F-5’-CTT CCT TTT ATT TTC CTT ACA GGG ATT-3’ and R-5’-GTA GGG ATG TAG ATT AAC CTT TTA TCT-3’ [20]. The PCR reaction consisted of heating for 5 min at 96℃ before being cycled through 35 cycles at 94℃ for 1 min, 56℃ for 1 min, 72℃ for 1 min and a final extension of 10 min at 72℃. After that, 10 units of HinfI enzyme were added to the PCR products, which would then be incubated at 37℃ for two to three hours. The post-incubation restriction enzyme digestion products were electrophoresed on a 3% agarose gel on multiSUB Choice, Wide Midi Horizontal Electrophoresis System (Cleaver Scientific, SKU: MSCHOICE10) for analysis.
Minisequencing: This technique was also used to detect the absence of exon 7 of the SMNt gene based on the exonic mismatch at position 214 in exon 7 (C in SMNt and T in SMNc). The primers for the technique had the following sequence as described in 2003 with some modifications [21]. First, the genetic region on exon 7 would be amplified at the annealing temperature at 54℃ with a pair of primers: F-5’-AGA CTA TCA ACT TAA TTT CTG ATC A-3’ and R-5’- CAC CTT CCT TCT TTT TGA TTT TGT-3’. Then, this segment would undergo the minisequencing reaction according to the instruction of the kit ABI PRISM Snapshot Multiplex with the primer: 5’-CCT TTT ATT TTC CTT ACA GGG TTT-3’. Later, the post-cycled products would be detected and analyzed by capillary electrophoresis on the 3130xl Genetic Analyzer. The minisequencing technique would eventually produce one (homozygous deletion of SMNt) or two (homozygous wild-type or heterozygous deletion of SMNt) peaks according to the sequence of this locus.
Prevention of DNA Contamination
Since SMA is a recessive disorder, polymorphic markers D5S629, D5S641, D5S1977 were evaluated to detect exogenous DNA contamination as previously reported in 2003 [15]. Haplotype analysis for all three microsatellites was performed to assess the possibility of contamination, especially in clinical PGT-M procedures.
Results
Lymphocyte and Blastomere Testing
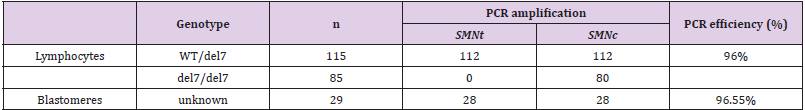
Single cells from SMA homozygotes and heterozygotes were used to test the accuracy and efficiency of the established method. The obtained results were annotated from electrophoresis and presented in Table 1. In 200 single lymphocytes collected, PCR amplicons were generated and observed on UV-laminated gel in 192 cells. Specifically, in 112 of 115 single lymphocytes with the heterozygous WT/del7 genotype (carrier), the expected PCR signal for exon 7 of the gene SMNt and exon 7 of the gene SMNc were observed (97.4%). 80 in a total of 85 lymphocytes isolated from SMA patients, only exon 7 of SMNc was observed after amplifying. Thereupon, since the genotypes of these single cells were known, the accuracy for this assay was 100%, and the overall PCR efficiency was 96%. Twenty-nine surplus embryos at the blastocyst stage were enrolled and tested (their results also were illustrated in Table 1).
The embryos’ origin came from couples without SMA history. Thus the genotype of blastomeres was unknown. In every sample, exon 7 of the SMNt and SMNc gene were expected, which allowed excluding the SMA homozygote circumstances. A positive amplification outcome was observed in 28/29 samples, whereas one case failed to amplify the target exon. Hence, based on the detected products, the amplification efficiency for this assay on invitro fertilized embryos was calculated to be 96.55%, approximate to that of lymphocytes. In conclusion, our protocol indicated the efficiency and reliability for distinguishing the exon 7 of pathogenic gene SMNt from its centromeric copy; moreover, it can be offered to establish a novel PGT-M procedure for SMA.
Clinical PGT-M of SMA with PCR-RFLP
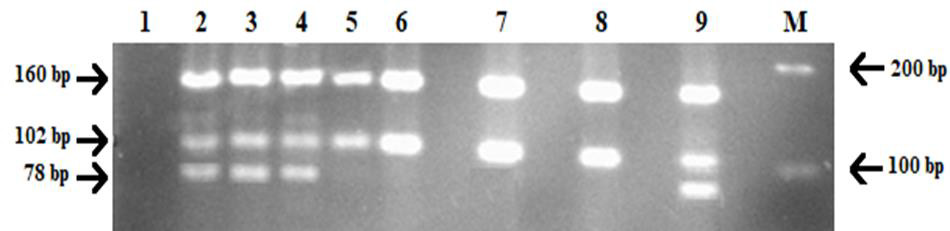
A Vietnamese 42-year-old and 38-year-old couple (coded SMA18) previously had a child who was diagnosed with spinal muscular atrophy at one year old. The child’s symptoms and mutation were discovered by the Vietnam National Hospital of Pediatrics, the identified cause was SMA with the homozygous absence of exon 7 of the SMNt gene. Ovarian stimulation, oocyte retrieval, and ICSI were performed using standard protocols, their 4 blastocysts were opened ZP and aspirated blastomeres followed by applying with the same genetic analysis that was previously tested above. The PCR-RFLP assay was conducted to diagnose spinal muscular atrophy on these samples and the obtained results are shown in Figure 1. Based on the gel separation result, embryo numbers 1, 2, and 3 showed the same pattern as the electrophoresis products of SMA patients. Consequently, they were determined to be mutation homozygotes. Under the UV light, the bands appeared bright at the corresponding position 160 bp and 102 bp, lacking the 78 bp segment of SMNt. However, at lane 9, the restriction enzyme products of embryo no.4 were separated into three different bands at the approximate size as the positive control – having at most one mutant allele. Therefore, the first three embryos were diagnosed to be all affected with SMA due to the exon 7 deletion on the survival motor neuron gene while the ladder was not.
Figure 1: Results of PGT-M of SMA18 using PCR-RFLP with HinfI restriction enzyme digestion.
• Lane 1: PCR negative control (DNA blank)
• Lane 2: PCR positive control (genomic DNA)
• Lane 3, 4: SMA carriers (SMA18 parents)
• Lane 5: SMA homozygous deletion of exon 7 of SMNt (SMA18 first born)
• Lane 6, 7, 8: embryo no.1, 2, 3
• Lane 9: embryo no.4.
Clinical PGT-M with Mini Sequencing
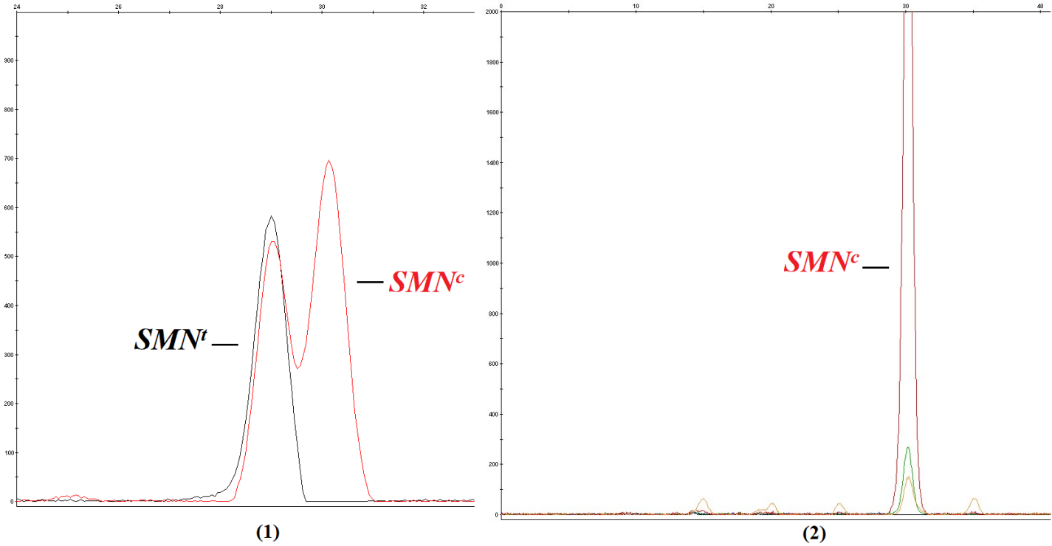
In addition to PCR-RFLP, we also conducted the minisequencing technique in PGT-M of SMA disease for the family SMA18 and received consistent results. The obtained electropherograms for the diagnosis technique were shown in Figure 2. By annotating the results of minisequencing, only embryo no.4 presented two peaks corresponding to exon 7 of the gene SMNt and exon 7 of the gene SMNc. In contrast, the remaining three embryos demonstrated only one peak corresponding to exon 7 of the gene SMNc. Accordingly, the PGT-M results of both techniques were identical; they all indicated that only embryo no.4 among four embryos was unaffected with the disease despite having the possibility of carrying one mutated allele.
Figure 2: Results of PGT-M of SMA18 using minisequencing.
1. Minisequencing result of embryo number 4, the graph showed two peaks (in black and in red) corresponding to both SMNt and SMNc – the pattern for homozygous wildtype or heterozygous genotype
2. Minisequencing result of embryo numbers 1, 2, 3 showed only one peak (in red) corresponding to only SMNc – the homozygous deletion pattern.
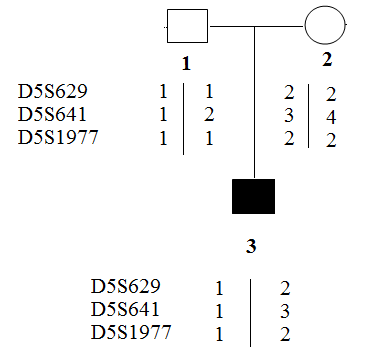
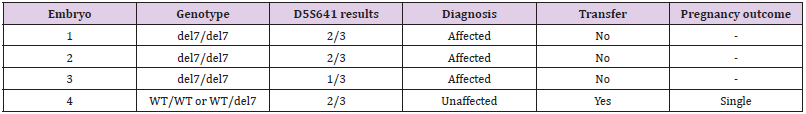
Assessing Exogenous DNA Contamination in SMA18 PGT-M Procedure: Polymorphic markers as D5S629, D5S641 and D5S1977 were amplified on the DNA samples of the SMA18 couple to assess the contamination state of the DNA sample. The obtained results were illustrated in Figure 3. Of the three markers tested, only marker D5S641 was semi-informative, while the others were not. Hence, D5S641 was used to assess exogenous DNA contamination in PGT-M for the SMA18 family. The complete summary of the results containing the overall genotypes, amplification of polymorphic markers, and pregnancy outcome of this family were described in Table 2. The only unaffected embryo was transferred into the mother’s uterus and then, luckily achieved pregnancy followed by a baby born. The neonatal genetic diagnosis was performed and confirmed that the child was free of SMA.
Figure 3: Results of PCR of polymorphic markers.
1. Father of SMA18 patient
2. Mother of SMA18 patient
3. SMA18 patient.
Discussion
The finding and characterization of the SMN gene as the candidate gene for SMA, reported in 1995, has provided tools to perform mutation analysis in the affected patients and the inheritance in their families [21]. In particular, the homologous sequence SMNc on the gene allows the discrimination between a deletion and PCR failure and helps prevent misdiagnosis by serving as an internal control for the PCR reaction. In the same year, another paper was published on a PCR-based protocol tailored for the exon 7 of the SMNt gene, in which a restriction enzyme was used to discriminate between the gene and its highly homologous copy sequence [22]. Theoretically, couples with SMA-affected children have a 25% risk of having a child with the disease; therefore, PGT-M should be considered regarding the possibility of having healthy offspring. In PGT-M, unaffected embryos would be selected before implantation by genetic analysis. Some protocols are available for the preimplantation diagnosis of SMA. Most of them consist of two rounds of PCR to amplify the exon 7 of the SMNt gene directly from a single cell, followed by enzymatic digestion. These researches produced a similar outcome with a PCR efficiency of around 90 - 95% [16,20,23].
In 2003, the diagnosis for the disease was carried out with a single round PCR strategy by utilizing allele-specific primers, then resulted in a procedure with the efficiency reaching nearly 99% [15]. Another protocol was to detect the presence of exon 7 of the SMNt gene by using the minisequencing method, which was conducted in 2003 with an efficiency of 96% [24]. However, the essential characteristics in employing these methods may lead to misdiagnosis as amplification failure (allele drop out - ADO) and contamination. Therefore, our strategy was to conduct genomewide techniques (Whole Genome Amplification) with the genome amplified thousands of times before proceeding to the subsequent analysis and amplify the polymorphic markers to detect exogenous DNA contamination. According to prior studies on WGA, the multiple displacement technique (MDA) – the main principle of the REPLI-g® Single Cell Kit was a robust, reliable, and accurate tool for genomic analysis. Thanks to the utilization of the Phi 29 DNA polymerase and random exonuclease-resistant primers, the highfidelity DNA yields collected from biopsied cells were significantly improved compared to PCR-based amplifying methods.
Based on the data provided by the manufacturer, the amplified DNA was more than 10 kb in length and uniform across the whole genome. Moreover, various downstream assays could be performed simultaneously with the MDA products, such as genotyping single nucleotide polymorphisms, RFLP analysis, and sequencing [25- 27]. Therefore, with the application of WGA at the first step of preimplantation genetic diagnosis, our research overcame the limits of low DNA quantity in single cells and ensured that the output DNA covering the whole genome was sufficient to be utilized in the diagnostic process. With the WGA reaction, PCR efficiency for the detection of exon 7 in lymphocytes and embryonic cells reached 96% and 96.55%, respectively. Compared to the studies previously published worldwide, the primers for polymerase chain reaction were slightly modified. The reaction’s conditions were also optimized to reduce the amplification failure and ADO rate, resulting in a consistent figure in PCR efficiency. However, the diagnosis of SMA at the preimplantation stage might encounter the situation where false-negative results were produced due to contaminations [28]. Hence, incorporating polymorphic markers to reduce exogenous DNA contaminants was crucial to the PGT-M procedure of any monogenic recessive disease. Introduced in a duplex PCR in 2003, the importance of these short tandem repeat markers in diagnosis assay was undeniable and widely recognized [15,18,23].
Consequently, to avoid misdiagnosis which unfortunately happened before in a PGT-M for SMA without linked polymorphic markers, our primary approach was to integrate different techniques to diminish the possible shortcomings contributing to a false result [29]. Furthermore, setting up a novel protocol for the monogenic disorder is work-intensive. The procedure needs to improve the accuracy and efficiency of genetic analysis and embryo biopsies. Compared to cleavage-stage biopsy with one aspirated cell, trophectoderm biopsy provides more blastomeres for DNA amplification. As a result, the ADO rate of TE biopsy were 9.8% lower than that in the other method were 15.3% [18]. Scott Jr. et al. performed a paired RTC; they reported that although cleavagestage biopsy markedly diminished viability and implantation potential, TE biopsy might be safe and had no measurable impact when indicating PGT-M [30]. A persuasive explanation for this difference is that a smaller proportion of aspirated blastomeres in total cellular constitution of blastocyst than that in cleavage embryo and avoiding inner cell mass perhaps more tolerant to manipulation. In addition, the new TE biopsy modification opening ZP on blastocyst stage was applied; it was suitable for both hatching as well as non-hatching embryos, decreased extra stress caused by prolonged exposure to the suboptimal embryo culture medium, and avoided the side effect of rising temperature following laser shooting at the cleavage-stage ZP opening [30-33].
Conclusion
The established protocol for preimplantation genetic diagnosis of spinal muscular atrophy was helpful for carrier couples in Vietnam as well as in other countries to get healthy children. The application of the protocol in clinical setting would benefit families with SMA with accurate and reliable results so that they would receive appropriate genetic consultations depending on their needs.



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.