Molecular Docking, Synthesis, in Silico and in vitro Screening of Substituted Aryl Ureido Analogues as BACE1 Inhibitors to target Alzheimer’s Disease
Introduction
In Alzheimer’s disease, one of the defining crucial hallmark in brain is the formation of extracellular amyloid-beta (Aβ) peptide plaques and the other being intracellular aggregation of hyper phosphorylated tau to form neurofibrillary tangles. Literature directs, cerebral Aβ accumulation begins at an earlier stage i.e. 10-20 years before the inception of dementia, which indicates that targeting Aβ deposition in brain will be apt for AD pathogenesis [1]. BACE1 (β-site APP cleaving enzyme, β-secretase) is the first protease of the amyloid cascade which initiates cleavage of the amyloid precursor protein (APP) to generate two fragments viz. N terminal fragment and a Aβ-containing C-terminal fragment. Subsequently, the C-terminal fragment is cleaved by γ-secretase to liberate Aβ peptides (Aβ40 or Aβ42) and an APP intracellular domain (AICD). In AD, Aβ42 is over produced due to genetic mutations of either the APP gene or other genes. The Aβ42 peptide is more hydrophobic and stickier than Aβ40 so it readily clumps resulting in formation of amyloid plaques [2,3].
As BACE1 is the key rate limiting enzyme of the amyloidogenic pathway so, inhibition of β-secretase can be considered as a prominent therapeutic target for treatment of AD as the disease still lacks an effective treatment [4-6]. BACE1 is highly expressed in brain and belongs to aspartyl protease family [7]. Literature survey indicates that the active site of BACE1 has two catalytic aspartic residues (Asp32 and Asp228),[8] optimum acidic pH and the correct sequence specificity for enzymatic activity [5]. The reported crystal structure of BACE1 complexed with OM00-3, an octapeptide inhibitor (PDB: 1m4h); depicts eight sub sites for ligand-enzyme interaction [9]. Literature reports discovery of many peptidomimetic as well as non-peptide BACE1 inhibitors [10,11]. Although, peptidomimetic inhibitors displayed potent activity against BACE1, their relatively large molecular size, low metabolic stability and poor bioavailability rendered their development into therapeutic drug candidates difficult [12]. Hence, now the research interest are directed more towards development of small non-peptide organic moieties as lead compounds [13-16].
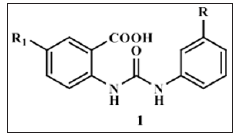
Some notable small-molecule nonpeptide BACE1 inhibitor classes are Vertex biarylnaphthalenes. acylcarbinamine, acylguanidine, aminothiazines, and aminoquinazoline-based scaffolds [6]. Huang and co-workers have reported their work on cell permeable diphenyl urea analogues [17]. Zhu et al revealed unique interaction pattern of CNS penetrant amidine containing heterocylic analogues with BACE1 catalytic dyad [18]. Prompted by above findings we tried to explore the possibility of substituted aryl groups on the two amidine nitrogen’s and replacement of one of the nitrogen with oxygen to simulate ureido functionality. By this approach we designed, synthesized and screened substituted aryl ureido analogues (1) derived from substituted anthranilic acid as potential non-peptide BACE1 inhibitors. In silico ADME studies were carried out to evaluate drug like properties of the candidates (Figure 1).
Results and Discussion
Docking approach was employed to explore the BACE1 active site and to estimate binding interactions of these analogues with the receptor. For docking studies, the protein data bank (PDB: 1m4h) crystal structure of BACE1 complexed with OM00-3 inhibitor possessing good potency (Ki: 0.3 nM) was considered. This inhibitor exhibits significant binding to the sub sites and has been extensively studied [19]. The docking protocol was validated by reproduction of binding pose of OM00-3 inhibitor in the 1m4h active site (rmsd 1.51). The octapeptide inhibitor OM00-3 of PDB 1m4h occupies S1, S2, S1ꞌ and S2ꞌ sub sites. To judge further, some reported non-peptide BACE1 inhibitors were docked in the BACE1 active site. Many exhibited interactions with the crucial catalytic dyad involved in drug-receptor binding and enzymatic catalysis. They occupied S1, S2 and S1ꞌ sub sites in the active site [20]. As per the Hong et al amino acid residues present in various sub sites were: S1- Leu30, Asp32, Tyr71, Gln73, Phe108, Asp228, Gly230; S2- of Tyr71, Thr72, Gln73, Gly230, Thr231, Arg235 residues and S1ꞌ- Gly34, Tyr71, Thr232 and Arg307 [9]. On the same guidelines, series of 2-(3-substitutedphenylureido)-5-nitrobenzoic acids (Type I) was docked in the BACE1 active site (Table 1).
These analogues were found to be oriented in the centre of BACE1 binding pocket, occupying S1, S2 and S1ꞌ sub sites analogous to the reported docked inhibitors. In Type I series, the 5-nitro group in all analogues exhibited bidentate interactions (H-bond and electrostatic) with Gln73 residue through water molecule except 4c wherein the nitro group interacted with Thr72. Interestingly, in 4c both the ureido nitrogen’s formed H-bonds with the catalytic residue Asp32 of BACE1. In the other Type I analogues, apart from the nitro group, ureido as well the carboxylic acid group depicted interactions with BACE1. Like reported inhibitors, they were associated with Asp32, Gly34, Thr72 and Gly230 residues of BACE1 active site. These findings were similar to the reported motifs so it can be envisaged that these 2-(3ꞌ-(3ꞌꞌ-substitutedphenyl)ureido)-5-nitrobenzoic acids successfully bind to the BACE1 catalytic site and should therefore turn out to be fruitful candidates for BACE1 inhibition (Figure 2).
Further, to study the effect of introduction of basic group on interaction pattern of Type I series within BACE1 active site, the acidic nitro group was replaced by amino group to obtain a new series of 5-(3ꞌ-(3ꞌꞌ-substitutedphenyl)ureido)-2-aminobenzoic acids (Type II) (Table 2). These analogues occupied S1 and S1ꞌ sub sites. Analogous to 4c, nitrogens of the ureido group of all analogues of this new series depicted bidentate interaction with Asp32 via H-bond and electrostatic interactions except 5f whose ureido group interactions where similar to other Type I members. The newly introduced 2-amino group formed H-bonds either directly or through water molecule with Lys107 and Phe108 residues, which was different from Type I series ligands. On superimposing these Type II analogues with the Type I ligands, orientation of both the series was found to be different which may account for change in the interaction pattern within BACE1 active site (Figure 3).
In Silico ADME Studies
The in silico ADME (Absorption, Distribution, Metabolism and Excretion) properties of designed test set analogues and reported ones were determined to gain an understanding of their pharmacokinetic behavior invivo. The 2-(3ꞌ-(3ꞌꞌ-substitutedphenyl)ureido)-5- nitrobenzoic acids (Table 1) possessed descriptor values within the recommended range of QikProp software such as low molecular weight (MW < 350 g/mol), hydrogen bond donor (HBD) < 3 and QP logPo/w values in the range of 1.0-2.4 [21]. Bioavailability can be gauged by estimating the QP logP values, literature states that for therapeutic use, compounds with QP logP values lower than 5 may have improved drug properties compared to more hydrophobic analogues [22]. The QP logP values for these analogues appear to be favourable. Ghose et al has mentioned some guidelines to design good quality CNS drugs: favorable QikProp CNS parameter, total solvent accessible surface area (SASA) of 460−580 Å2 and HBD less than 3 [23]. For permeability through the blood brain barrier (BBB), QikProp predicts QP logBB; the brain-blood partition coefficient. The QP logBB values were obtained in range of -1.9 to -2.7 (Expected range: -3-1.2) except 4f analogue.
Similarly, the SASA values were found to be in the range of 572- 610, very much favourable to be the likely CNS candidates. Series of 5-(3ꞌ-(3ꞌꞌ-substitutedphenyl)ureido)-2-aminobenzoic acids (Table 2) exhibited some values similar to Type I series such as low molecular weight (MW < 320 g/mol). Comparison of QP logPo/w values (0.5-2.0) with Type I analogues indicated that they are less hydrophobic and HBD value was 4, well within QikProp limit but a digit more than the limit specified by Ghose et al. SASA values were observed to be in between 574-600, seems to be favourable. QP logBB values were observed in the expected range to be-1.6 to -2.9. The percent Human oral absorption for both the series were found to be moderate i.e. 40-60%. None of them violated the Lipinski’s rule of five.
Synthesis
The Type I series docking results were analogous to reported ones so the analogues were synthesized and the structures were confirmed by use of spectral techniques (Scheme 1). In addition, the in silico ADME results were more favourable for the Type I analogues. Further, they were subjected to screening for activity against BACE1.
Pharmacological screening
The in vitro BACE1 FRET (Fluorescence resonance energy transfer) inhibition study results were in agreement to the molecular docking outcome. All analogues of the series 2-(3ꞌ-(3ꞌꞌ-substitutedphenyl) ureido)-5-nitrobenzoic acids showed BACE1 enzyme inhibition at 50μM concentration (Table 1). The percent inhibition at 50μM concentration was found to be in the range of 20-40 %.
Conclusion
The intention to design this scaffold was to analyse the suitability of carboxylic acid group possessing compounds as prospective CNS candidates; as literature indicates that carboxylic acid group is not a very common functionality in the design of CNS drugs. It has been observed that many non-CNS drugs especially orally administered ones have carboxylic acid group while very few CNS drugs possess it. Only 3−4% of CNS drugs had a carboxylic acid group, whereas 25% of non-CNS oral drugs possessed a carboxylic acid group. The docking and in vitro screening studies comply with our assumptions that the substituted aryl ureido analogues can be targeted for potential CNS use. Comparison between the two serie projects the 2-(3ꞌ-(3ꞌ substitutedphenyl)ureido)-5-nitrobenzoic acids (Type I) to be more suitable candidates than Type II series. Amongst them the 4c and 4d analogues exhibit equivalent BACE1 enzyme inhibition. These analogues can be further optimized by suitable structural modifications to attain a more targeted concentration at the site for higher favourable response as BACE1 inhibitors.
Experimental Section
Molecular Docking
Materials: On Red Hat Enterprise Linux (RHEL) workstation various software’s viz. Sybyl version 8.1.1 (Tripos International, Portugal) [24] and GOLD version 4.1.1 (CCDC Ltd., UK) [25] were utilized for ligand as well as protein preparation and docking studies. Designing of ligands and protein preparation. The method already reported in our publication was followed for ligand and protein preparation. The same protein data bank crystal structure (PDB code 1m4h) in which BACE1 is complexed with an octapeptide OM00-3 was prepared and further considered for docking studies [26].
Designing of ligands and protein preparation: The method already reported in our publication was followed for ligand and protein preparation. The same protein data bank crystal structure (PDB code 1m4h) in which BACE1 is complexed with an octapeptide OM00-3 was prepared and further considered for docking studies [26].
Docking protocol: The earlier published protocol was followed for docking studies of these analogues [26].
In silico ADME studies
Materials: For the studies Qik Prop version 3.0 (Schrodinger LLC, New York, USA) software on RHEL workstation was employed [21].
Method: Same method as followed in our earlier work was utilized to determine various descriptor values for the in silico ADME studies [26].
Synthesis
Materials: As per the requirement solvent used were distilled for purification purpose. Merck
F254 TLC plates were used to monitor progress of reaction. The title compounds were purified using recrystallization and/ or column chromatographic technique. Infrared (IR) spectra were recorded on Shimadzu IR Affinity-1 FTIR spectrophotometer using DRS-8000 (Diffuse Reflectance attachment). 1HNMR spectra and Mass spectra have been recorded on Mercury Plus 300 MHz (Varian, USA) NMR spectrometer and 410 Prostar Binary LC with 500 MS IT PDA detectors (Varian Inc, USA) mass spectrometer respectively.
For 1HNMR spectra tetramethylsilane (TMS) has been used as an internal standard and DMSO-d6 (Dimethyl sulfoxide) as solvent. 2-(3ꞌ-(3ꞌꞌ-Substitutedphenyl)Ureido)-5-Nitrobenzoic Acids (4 (a-f)): The 2-(3ꞌ-(3ꞌꞌ-substitutedphenyl)ureido)-5-nitrobenzoic acids were synthesized as per Scheme 1.
General Method for Analogues (4 (a-f)): To the solution of 2-amino-5-nitrobenzoic acid (2, 0.2 g, 1.0989 mmol) in 2 ml dry acetone, equimolar solution of respective 3-substitutedphenyl isocyanate (3 (a-f)) in 2 ml dry acetone was added gradually at 40oC in 15-20 minutes with stirring. The reaction mixture was refluxed at 40oC for 16 hours with addition of minimum amount of dry acetone if needed. Further, mixture was poured in 50 ml water for precipitation of crude product which was recrystallized from ethanol resulting in pure analogues as product (5 (a-f)) [27].
2-(3ꞌ-(3ꞌꞌ-acetylphenyl)ureido)-5-nitrobenzoic acid (5a): Yield: 64%; 206-08oC. IR (KBr) ʋ: 1708 (ureido), 3089 (C-H), 1695 (>C=O), 1593, 1357 (NO2), 1681 (COCH3) cm-1. 1HNMR (DMSO-d6): δ 10.60 (s, 1H), 10.28 (s, 1H), 9.33 (s, 1H), 8.19 (d, 1H, J = 8.8 Hz), 8.12 (t, 1H, J = 1.6 Hz), 7.90 (d, 1H, J = 8.8 Hz), 7.85 (d, 1H, J = 2.6 Hz), 7.82 (td, 1H, J = 6.6, 2.3 Hz), 7.63 (dd, 1H, J = 5.3, 2.4 Hz), 7.47 (t, 1H, J = 7.9 Hz), 2.57 (s, 3H). MS (acetone) [M]: 342.
2-(3ꞌ-(3ꞌꞌ-Chlorophenyl)Ureido)-5-Nitrobenzoic Acid (5b): Yield: 73%; 218oC. IR (KBr) ʋ: 1720 (ureido), 3035 (C-H), 1691 (>C=O), 1597, 1355 (NO2) cm-1. 1HNMR (DMSO-d6): δ 10.60 (s, 1H), 10.25 (s, 1H), 9.31 (s, 1H), 8.19 (d, 1H, J =8.8 Hz), 7.86 (d, 1H, J = 2.2 Hz), 7.83 (d, 1H, J = 2.2 Hz), 7.75 (t, 1H, J = 2.0 Hz), 7.40 (td,1H, J = 8.2, 1.6 Hz), 7.33 (t, 1H, J = 7.9 Hz), 7.08 (td, 1H, J = 5.7, 1.7 Hz). MS (acetone) [M]: 334.
2-(3ꞌ-(3ꞌꞌ-Cyanophenyl)Ureido)-5-Nitrobenzoic Acid (5c): Yield: 36%; 208oC. IR (KBr) ʋ: 1708 (ureido), 3055 (C-H), 1697 (>C=O), 1591, 1346 (NO2), 2249 (CN) cm-1. 1HNMR (DMSO-d6): δ 10.65 (s, 1H), 10.40 (s, 1H), 9.30 (s, 1H), 8.19 (d, 1H, J = 8.8 Hz), 8.00 (t, 1H, J = 6.6 Hz), 7.91 (d, 1H, J = 8.8 Hz), 7.85 (d, 1H, J = 2.2Hz), 7.75 (d, 1H, J = 8.1 Hz), 7.63 (d, 1H, J = 2.2 Hz), 7.48 (t, 1H, J = 6.2 Hz). MS (acetone) [M]: 325.
2-(3ꞌ-(3ꞌꞌ-Methoxyphenyl)Ureido)-5-Nitrobenzoic Acid (5d): Yield: 63%; 198oC. IR (KBr) ʋ: 1724 (ureido), 3034 (C-H), 1697 (>C=O), 1598, 1359 (NO2), 2843 (OCH3) cm-1. 1HNMR (DMSO- d6): δ 10.51 (s, 1H), 10.04 (s, 1H), 9.31 (s, 1H), 8.18 (d, 1H, J = 8.4 Hz), 7.83 (dd, 1H, J = 8.8, 2.6 Hz), 7.22 (t, 1H, J = 2.2 Hz), 7.21 (d, 1H, J= 1.1 Hz), 7.17 (d, 1H, J = 6.2 Hz), 7.08 (td, 1H, J = 8.1, 1.1 Hz), 6.61 (td, 1H, J = 8.1, 2.7 Hz), 3.74 (s, 3H). MS (acetone) [M]: 330.
2-(3ꞌ-(3ꞌꞌ-Methylthiophenyl)Ureido)-5-Nitrobenzoic Acid (5e): Yield: 63%; 184-186oC. IR (KBr) ʋ: 1720 (ureido), 3082 (CH), 1697 (>C=O), 1585, 1357 (NO2), 678 (SCH3) cm-1. 1HNMR (DMSO-d6): δ 10.54 (s, 1H), 10.07 (s, 1H), 9.31 (s, 1H), 8.18 (d, 1H, J = 8.8 Hz), 7.83 (dd, 1H, J = 8.6, 2.4 Hz), 7.50 (t, 1H, J = 1.8 Hz), 7.30 (d,1H, J = 1.8 Hz), 7.28 (t, 1H, J = 5.1 Hz), 7.25 (dd, 1H, J = 8.2, 3.5 Hz), 6.91 (td, 1H, J = 7.3, 1.5 Hz), 2.47 (s, 3H). MS (acetone) [M]: 346.
2-(3ꞌ-(3ꞌꞌ-Nitrophenyl)Ureido)-5-Nitrobenzoic Acid (5f): Yield: 74%; 202oC. IR (KBr) ʋ: 1720 (ureido), 3088 (C-H), 1703 (>C=O), 1602, 1354 (NO2) cm-1. 1HNMR (DMSO-d6): δ 10.70 (s, 1H), 10.57 (s, 1H), 9.32 (s, 1H), 8.56 (t, 1H, J =2.0 Hz), 8.19 (d, 1H, J = 8.8 Hz), 7.88 (d, 1H, J = 2.9 Hz), 7.85 (dd, 1H, J = 6.0, 2.7 Hz), 7.62 (t, 1H, J = 3.1 Hz), 7.59 (d, 1H, J = 7.3 Hz), 7.56 (d, 1H, J = 3.7 Hz). MS (acetone) [M]: 345.
Pharmacological Screening
Materials: BACE1 FRET (Fluorescence resonance energy transfer) inhibition study was performed by Department of Pharmaceutical Sciences, University of Bologna, Italy, by the M-2420 method [7,28].
Preliminary Screening: Similar procedure as reported in our earlier published work was followed for the BACE1 inhibition study using the same M-2420 method [26].
Lower Trapezius Muscle Transfer For Elbow Extension Reconstruction After Failed Nerve Transfer for Tetraplegia-https://biomedres01.blogspot.com/2020/08/lower-trapezius-muscle-transfer-for.html
More BJSTR Articles : https://biomedres01.blogspot.com




No comments:
Post a Comment
Note: Only a member of this blog may post a comment.