Insilico Molecular Docking of Some Isolated Selected Compounds of Phoenix Sylvestris (L.) for Analgesic Activity
Introduction
Pain is an unpleasant sensory and emotional experience associated with actual or potential tissue damage or described in terms of such damage [1]. Irritations of pain receptor are given a feeling of pain, which can be found in the skin, joints and many internal organs. The cause of pain may also be damage to the nervous system, both the peripheral nerves, brain, and spinal cord. Pain can also occur without damage to tissues, although the patient refers to it (psychogenic pain). The process of pain is a complex phenomenon [2]. In the brain, pain stimulus is processed, and generated impulses are sent down the spinal cord following the appropriate nerves and instruct the body to respond, for instance withdrawing your hand from the fire. Peripheral nerves transmit pain stimulus to the spinal cord which then links to the brain [3]. An analgesic or painkiller is any member of the group of drugs used to achieve analgesia, relief from pain. They are distinct from anesthetics, which reversibly eliminate sensation. Analgesic substances interact with the transmitters and modulators of the pain system are helpful for many people with pain, but there is a great need for the development of better methods for the alleviation and control of both acute (immediate) and chronic (long-term, pathological) pain [4].
Analgesics including Nonsteroidal Anti-Inflammatory Drugs (NSAIDs) are commonly prescribed group of drugs in clinical practice for the management of pain and inflammation [5,6]. They work by interfering with Cyclooxygenase [COX] pathway, which involves the conversion of arachidonic acid by the enzyme COX to prostaglandins [7,8]. To suppress pain, NSAIDs are prescribed all over the world [9]. For severe or chronic malignant pain uploads analgesics are drugs of choice [3]. Despite wide clinical use of classical NSAIDs as analgesics, antipyretics, and anti-inflammatory agents their gastrointestinal toxicity is a major clinical limitation [5]. Different kinds of Studies have shown that opiates cause physical dependency, tolerance, and addiction while NSAIDs usually cause gastrointestinal disorders [10]. In this regard, herbal medicines from medicinal plants have been employed in Complementary and Alternative Medicine (CAM) for the treatment of pain as well as diseases related to these conditions [11].
The use herbal medicine is gaining support and recognition across the world because most of these products are believed to have bioactive compounds responsible for healing various diseases without any side effects and at a lower cost. Phoenix Silvestris Roxb. (Arecaceae), locally known as Khejur, is a palm tree cultivated for its syrupy juice and edible fruit in Bangladesh [12]. Palm is one of the important horticultural crops in many countries [13]. In Bangladesh, Khejur palm is produced as a homestead crop; however, it grows naturally or is cultivated in fallow lands, around homesteads, farmland boundary and even in the marginal lands along the roads and canals [14]. Fruits of the plant are used to treat back pain, stomachache, toothache, headache, arthritis, pain of buttocks, fever, piles, nervous debility, and as nervine tonic, restorative, sedative in ethnomedicine [12,15]. The sap of Khejur palm is a good source of vitamins of the B group and contains, in addition, a variable amount of ascorbic acid [16], freshly harvested sap consists of sucrose around 10%, minimal invert sugar of <0.5% and a small amount of protein, gums, and minerals.
Computational simulations of drug-target interactions using in silico molecular docking and molecular dynamics approaches are commonly used for the rational design and screening of drugs [17]. Molecular docking has become a major computational method for the prediction of ligand-receptor interactions [18]. A productive docking strategy must have the ability to adequately envision the local ligand represent the receptor limiting site (i.e. to find the trial ligand geometry inside a particular resistance confine) and the related physical-compound sub-molecular affiliations [19-22]. Furthermore, the aim of the study to find the mechanism of action of the isolated compounds from Phoenix Silvestris was explored the analgesic activity by molecular docking analysis.
Materials and Methods
In silico Molecular Docking Protein Preparation
Three-dimensional crystal structure of COX-1 (PDB id: 2OYE) and COX-2 (PDB id: 6COX) was downloaded in pdb format from the protein data bank [23]. After that, the structure was prepared and refined using the Protein Preparation Wizard of Schrödinger- Maestro v10.1. Charges and bond orders were assigned, hydrogens were added to the heavy atoms, selenomethionines were converted to methionines and all waters were deleted. Using force field OPLS_2005, minimization was carried out setting maximum heavy atom RMSD (root-mean-square-deviation) to 0.30 Å.
Ligand Preparation
The structures of six major representative compounds i.e., Diethylnitrosamine (CID: 5921), 2, 3-Dihydro-3,5-dihydroxy-6- methyl-4H-pyran-4-one (CID: 119838), 4-methylcatechol (CID: 9958), 2,4-Di-tert-butyl phenol (CID: 7311), and Diethyl Phthalate (CID: 6781) were obtained from PubChem database. The ligands were prepared with LigPrep tool embedded in Maestro 2015, neutralized at pH 7.0±2.0 using Epik and minimized by force field OPLS_2005.
Receptor Grid Generation
Receptor grids were calculated for prepared proteins such that various ligand poses bind within the predicted active site during docking. In Glide, grids were generated keeping the default parameters of van der Waals scaling factor 1.00 and charge cutoff 0.25 subjected to OPLS 2005 force field. A cubic box of specific dimensions centered around the centroid of the active site residues (Reference ligand active site) was generated for the receptor. The bounding box was set to 14 Å × 14 Å × 14 Å for docking experiments.
Glide Standard Precision (SP) Ligand Docking
SP flexible ligand docking was carried out in Glide of Schrödinger-Maestro v 10.1 [24,25]. within which penalties were applied to non-cis/trans amide bonds. Van der Waals scaling factor and partial charge cutoff were selected to be 0.80 and 0.15, respectively for ligand atoms. Final scoring was performed on energy-minimized poses and displayed as Glide score. The bestdocked pose with lowest Glide score value was recorded for each ligand.
Results and Discussion
In silico Molecular Docking Analysis
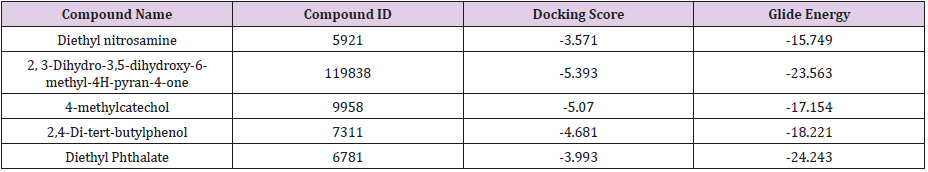
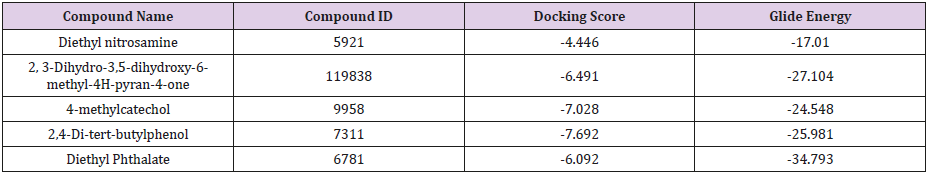
Prediction of interaction energies between ligand and receptor has been a major challenge for molecular docking [26]. Virtual screening utilizes docking and scoring of each compound from a dataset. The technique used is based on predicting the binding modes and binding affinities of each compound in the dataset by means of docking to an X-ray crystallographic structure [27]. Grid-based docking study was used to analyze the binding modes of molecules with the amino acids present in the active pocket of the protein [28]. To identify the potential analgesic lead molecule, we have subjected the docking analysis of the active compounds of Phoenix Silvestris to the active site of COX-1 and COX-2. In order to study the interaction of the selected compounds Diethyl nitrosamine, 2, 3-Dihydro-3,5-dihydroxy-6-methyl-4H-pyran-4- one, 4-methylcatechol, Diethyl Phthalate and 2,4-Di-tert-butyl phenol with COX-1 (PDB id: 2OYE) and COX-2 (PDB id: 6COX). We performed Glide docking analysis by Schrodinger suite v10.1, where among of these compounds 2, 3-Dihydro-3,5-dihydroxy-6- methyl-4H-pyran-4-one shows highest docking score against COX- 1enzyme and 2,4-Di-tert-butyl phenol shows highest docking score against COX-2 enzyme.
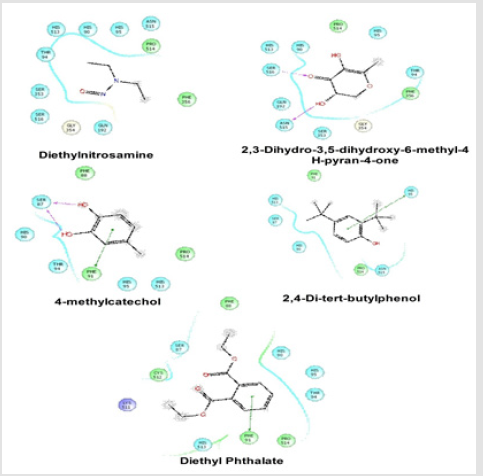
Docking Score suggested that 2, 3-Dihydro-3,5-dihydroxy- 6-methyl-4H-pyran-4-one and 2,4-Di-tert-butyl phenol had the highest affinity toward the COX-1enzyme and COX-2 enzyme corresponding to the other compound. The results of docking analysis were described in Tables 1 & 2 and the docking figure showed in Figures 1 & 2. The colors indicate the residue (or species) type: Red-acidic (Asp, Glu), Green-hydrophobic (Ala, Val,Ile, Leu, Tyr, Phe, Trp, Met, Cys, Pro), Purple-basic (Hip, Lys, Arg), Blue-polar (Ser, Thr, Gln, Asn, His, Hie, Hid), Light gray-other (Gly, water), and Darker gray-metal atoms. Interactions with the protein are marked with lines between ligand atoms and protein residues: Solid pink—Hbonds to the protein backbone, Dotted pink-H-bonds to protein side chains, Green—pi-pi stacking interactions, Orange-pi-cation interactions. Ligand atoms that are exposed to solvent are marked with gray spheres. The protein “pocket” is displayed with a line around the ligand, colored with the color of the nearest protein residue.
Figure 1: Schematic representation of the interactions between the best pose found for Diethyl nitrosamine, 2, 3-Dihydro-3,5- dihydroxy-6-methyl-4H-pyran-4-one, 4-methylcatechol, 2,4-Di-tert-butyl phenol and Diethyl Phthalate with COX-1 (PDB id: 2OYE).
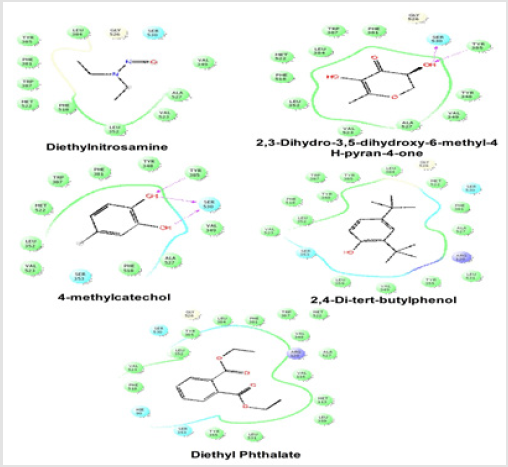
Figure 2: Schematic representation of the interactions between the best pose found for Diethyl nitrosamine, 2, 3-Dihydro-3,5- dihydroxy-6-methyl-4H-pyran-4-one, 4-methylcatechol, 2,4-Di-tert-butyl phenol and Diethyl Phthalate with COX-2 (PDB id: 6COX).
The gap in the line shows the opening of the pocket. The colors indicate the residue (or species) type: Red-acidic (Asp, Glu), Green-hydrophobic (Ala, Val,Ile, Leu, Tyr, Phe, Trp, Met, Cys, Pro), Purple-basic (Hip, Lys, Arg), Blue-polar (Ser, Thr, Gln, Asn, His, Hie, Hid), Light gray-other (Gly, water), and Darker gray-metal atoms. Interactions with the protein are marked with lines between ligand atoms and protein residues: Solid pink-H-bonds to the protein backbone, Dotted pink-H-bonds to protein side chains, Green pi-pi stacking interactions, Orange-pi-cation interactions. Ligand atoms that are exposed to solvent are marked with gray spheres. The protein “pocket” is displayed with a line around the ligand, colored with the color of the nearest protein residue. The gap in the line shows the opening of the pocket.
Conclusion
The present study revealed that between the compounds 2, 3-Dihydro-3,5-dihydroxy-6-methyl-4H-pyran-4-one and 2,4-Ditert- butyl phenol showed the best docking score against COX-1 and COX-2 respectively. Further, in vivo investigation needs to identify COX-1 and COX-2 enzyme inhibitory activity of isolated compounds from P. sylvestris.
Acknowledgment
The authors thankful Mr. Arkajyoti Paul for providing the software and helping in manuscript writing.
More BJSTR Articles: https://biomedres01.blogspot.com/



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.