MCLON3 and LMCD1 May Play a Vital Role in Young Colorectal Cancer with Poor Prognosis
Introduction
Background
Colorectal cancer is one of the most common malignancies in digest organs. Patients may suffer from this problem in all ages but be more inclined to happen after 50 years’ old. However, there is a tendency that colorectal cancer rising in the young group. What’s more, many young patients got a disappointed outcome eventually though had received radical operations, chemotherapies or many other active therapeutic treatments. Situation may be more complex that in some cases, young patients have a much poorer prognosis than aged patients even with an earlier stage or a higher differentiated type. However, in other cases, parts of young patients have an excellent outcome after operation. As is known, the deficiency of mmr is suggested as a cause of colorectal cancer. The status of cimp and cin are also considered important to the formation and development of cancer. The mutation of tp53, kras and braf have an impact on the prognosis and indict therapeutic targets. Therefore, we want to pursue something to this problem that is there any signature or molecular mark can we apply for predicting the outcome and becoming potential therapeutic targets? In order to settle this problem, we studied deep into the database of GSE40967 on the platform of GPL570, which includes 585 samples and complete clinical and pathological information.
Material and Method
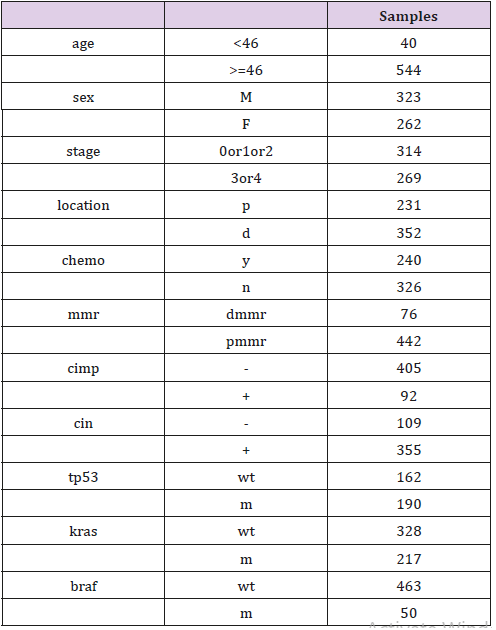
Ethics statement. The databases searched in our study are available online. Anyone is permitted to use all the data in the website of Array Express (http://www.ebi.ac.uk/arrayexpress/), which contains more than 65 thousands of experiments and about 2 millions of assays. It also supports for a search facility to get what type of array and the related information we want. All the gene expression omnibus datasets are also accessible to public from the GEO website (www.ncbi.nlm.nih.gov/geo/). Selection of databases. We searched in the Array Express with the condition of colorectal adenocarcinoma, filtered by organism of homo sapiens, experiment type of array assay and rna assay. As a result, 479 experiments satisfied were searched out. According to the data-integrity and sample quantity, we focus on the datasets of GSE40967 on the platform of GPL570, which contains 585 samples and records the details of age, sex, tumor location, stage, chemotherapy, overall survival(os), disease free survival(dfs), mmr, cimp, cin, tp53, kras and braf. Information of this dataset can be reviewed in Table 1.
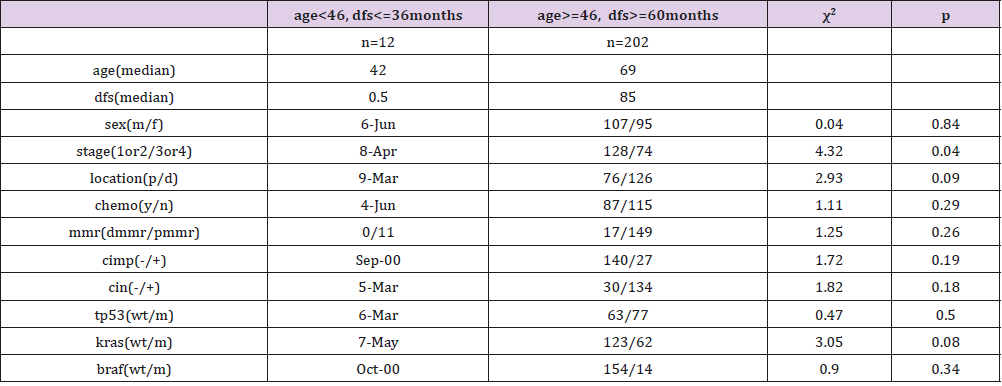
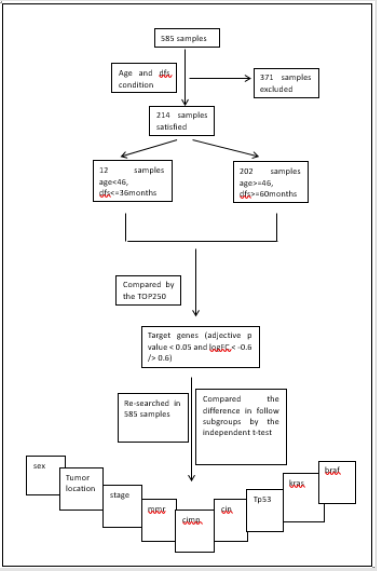
Identification of target specific genes. Although the sample quantity of this dataset is large, the group satisfying the condition of younger than 46 years and dfs no more than 36 months together only has 12 samples, and the control group satisfying the condition of equal to or older than 46 years and dfs no less than 60 months together has 202 samples. Those whose follow-up is not up to 36 months but still alive in the former group or not up to 60 months in the latter would be excluded. The details of two groups can be reviewed in Table 2. The genes of the two groups were then compared by the tool of GEO2R, TOP250, supplied in web of NCBI, which performs comparisons on original submitter-supplied processed data tables using the GEO query and limma R packages from the Bioconductor project and shows the top250 genes in difference. The genes would be selected out for adjective p value less than 0.05 and logFC less than -0.6 or more than 0.6. Then these genes would be taken into a further investigation. Association of target genes with key information in dataset GSE40967. The expression of the four genes was re-searched for all the 585 samples, and then their values were documented.
Every sample has recorded whole or parts of details of age, sex, tumor location, stage, chemotherapy, os, dfs, mmr, cimp, cin, tp53, kras and braf. And then the difference of the four genes’ expression was compared according to subgroups of sex, tumor location, stage, mmr, cimp, cin, tp53, kras and braf by two-independent-samplestest( p<0.05) in the software of spss16.0. Subgroups of nine valuables. To compare the difference of the four genes’ expression, sex subgroups were classified as male or female group, tumor locations were classified as proximal and distal colorectal cancer, stages were classified as 1/2 and 3/4, mmr status were classified as dmmr(mmr-deficient) and pmmr(mmr-proficient), cimp and cin status were classified as “–”and “+” respectively, and mutations of tp53, kras and braf were classified as mutation and wild type respectively. The design of this study shows as in Chart 1.
Results
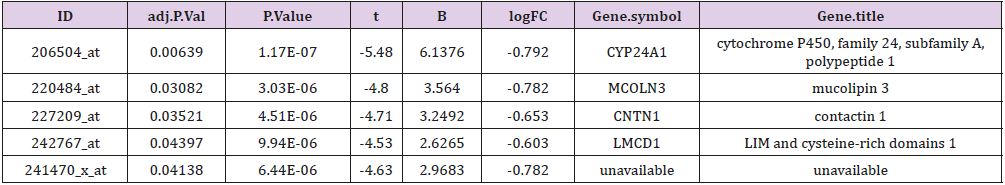
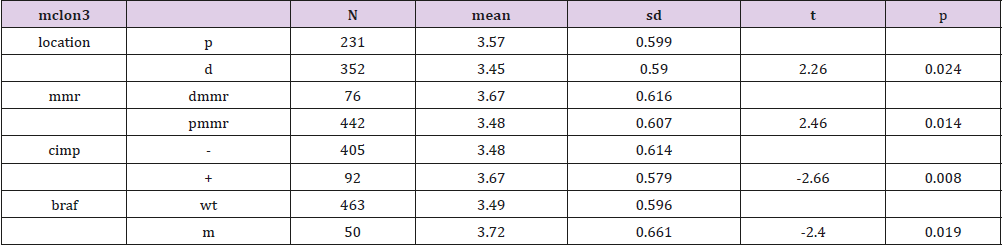
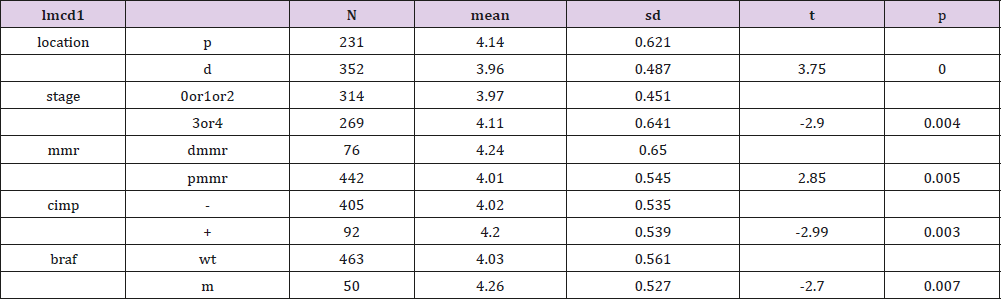
Genes of the two groups were compared by TOP250, and those with adj.p.val smaller than 0.05 and the magnitude of logFC bigger than 0.6 would be selected out. Five genes were found upregulated in the group of patients younger than 46 and with dfs less than 36 months, one of which was not available of the gene symbol or title, and the other four were cytochrome P450, family 24, subfamily A, polypeptide 1(CYP24A1), mucolipin 3(MCOLN3), contactin 1(CNTN1) and LIM and cysteine-rich domains 1(LMCD1) respectively (Table 3). After a re-comparison of the four available genes’ expression, including CYP24A1, MCOLN3, CNTN1, and LMCD1, in the different condition of sex, tumor location, stage, mmr, cimp, cin, Tp53, kras and braf in the 585 samples of the database of GSE40967 on the platform of GPL570, MCOLN3 and LMCD1 were thought to be two special genes. The MCOLN3 gene was found to be over-expressed in the proximal colon cancers (p=0.024), and in the status of mmr-deficient (p=0.014), cimp positive (p=0.008) and mutant type of braf (p=0.019) (Table 4). The LMCD1 gene was found to be over-expressed in the proximal colon cancers (p=0.000), in the TNM stage of 3 or 4 (p=0.004), and in the status of mmr-deficient (p=0.005), cimp positive (p=0.003) and mutant type of braf (p=0.007) (Table 5).
Discussion
The colorectal cancer occurs usually over 55 years old. In recent years, the incidence of colorectal cancer has become much higher in the young people with poorer prognosis. This phenomenon has been recognized but haven’t be illustrated well until now. Some factors in the young related to several known molecular mechanisms may explain that. MMR (mismatch repair), correcting mismatched nucleotides and insertion-deletion loops in DNA, is one important signal way [1,2]. CIMP (CpGislandmethylatephenotype) is associated with key factors related to worse disease outcome. Several studies had proved CIMP positive MSS BRAF mutated tumors living a particularly bad prognosis [3-8]. 10–20% of CRCs, have been found CIMP-positive associated with BRAF mutations [9]. CIN (chromosomal instability), which is characterized by structural or numerical chromosomal alterations, is detected in 85% of CRCs [10]. CIN can induce cancer development and progression and is associated with poor prognosis and drug resistance in CRC [11- 14]. Other molecular events, such as hypermethylation phenotype (CIMP+) [15,16], kras, braf and tp53 also have been thought as key impacts on CRC.
In order to find genes intensively related to colorectal cancer in young adult, we took advantage of datasets of GSE40967 on the platform of GPL570, which contains 585 samples and records the details of age, sex, tumor location, stage, chemotherapy, overall survival, disease free survival, mmr, cimp, cin, tp53, kras and braf. CYP24A1, MCOLN3, CNTN1 and LMCD1 were statistically thought to be special in the young patients. What’s more, MCOLN3 and LMCD1 were relevant with some important molecular events in colorectal cancer and may play a role in this process. The expression of the both genes is much higher in the event of dmmr, cimp mutant and braf mutant. In which, the expression of LCMD1 accelerates in the stage of III or IV. In human, the MCOLN3 gene resides on the short arm of chromosome 1 at 1p22.3. The encoded protein, TRPML3, has 553 amino acid with a predicted molecular weight of ≈64 kDa. TRPML3 is an inwardly-rectifying cation channel [17], and one of the largest families of cation channels [18]. TRP channels are involved in regulating various cellular functions, ranging from pure sensory function to molecular regulation, hence they serve as gatekeepers for transcellular transport of sodium and calcium ions [19,20]. TRPML3 is involved in autophagy regulation [21,22].
As known, cancer progression is closely related to dysregulations of the cell cycle and are accompanied by enhanced cell proliferation and/or suppression of apoptosis leading to cell death [23]. In contrast, degenerative processes are initiated by either enhanced cell death and/or suppression of cell proliferation. The key factors between these opposing biological processes often lead to a perturbed balance between the processes of proliferation, autophagy, and apoptosis [24]. Ca2+is one of the most important regulators of cell survival/death processes. As a second messenger, Ca2+ is able to activate or inactivate various regulatory proteins such as enzymes, transcriptional factors, or molecular chaperones. Importantly, Ca2+signaling has been shown to regulate cellular processes such as cell proliferation, survival, migration, invasion, motility, autophagy and apoptosis [25-27]. Recent studies by Kim et al. showed that over expression of TRPML3 leads to increased autophagy in HeLa cells and that TRPML3 channels are engaged to autophagosomes upon induction of autophagy [28]. Heteromultimerization of TRPML channels also have been shown to have a role in autophagy [29].
A recent study showed that TRPML3 interacts with mammalian ATG8 homologue GATE16 to regulate autophagy, thereby suggesting a vital role of TRPML3 in autophagasome maturation through the interaction with GATE16 [30]. LMCD1,LIM and cysteine rich domains 1, encodes a member of the LIM-domain family of zinc finger proteins [31]. The protein may act as a co-regulator of transcription along with other transcription factors. In other hand, LMCD1 is a novel pathway to upregulate Rac1 signaling, enhanced lamellipodia extension and cellmobility in vitro and in vivo [32]. Upregulation of LMCD1 through amplification is a necessary factor for cell migration, which has been found in HCC cells. Somatic alterations of LMCD1 have important roles in HCC tumor progression especially tumor cell migration and metastasis [33]. Mutant LMCD1 is a putative HCC proto-oncogene involved in dysregulated Rac1 signaling, which results in actin cytoskeleton re-organization and facilitates cell migration and metastasis. In summary, this study finds four genes, CYP24A1, MCOLN3, CNTN1 and LMCD1, related to the young CRC patients with poor prognosis. Especially the MCOLN3 and LMCD1 are tightly related to some unknown molecular pathway in tumorgenesis and progression of CRC, which may prove further diagnostic or therapeutic targets.
For more Articles: https://biomedres01.blogspot.com/



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.