In Silico Structure-Based Screening of Large Ligand Library Against Virulence Factors of Drug Resistant Pathogens
Introduction
The discovery and continual development of antibiotics since the 1940s have greatly reduced the mortality rate of infectious diseases. However, newly emerging and reemerging of drug-resistant microbes mean that despite our best efforts, infectious diseases remain a great threat to humanity today [1]. Among these, Staphylococcus Aureus and Mycobacterium tuberculosis (MTB) are especially problematic. S. aureus is a highly adaptive gram-positive bacterium and commensal of the human skin and nostrils [2]. S. aureus is a common cause of minor skin and wound infections, but can also cause serious and even fatal infections, particularly in the immunocompromised persons. Methicillin-resistant Staphylococcus Aureus (MRSA) infection is becoming an alarming local and global threat, in particular the number of both hospital-acquired and community-acquired cases are rising, leading to many life-threatening diseases such as endocarditis, pneumonia and toxic shock syndrome [3]. MRSA has been endemic in Hong Kong since the mid-1980s. Approximately 70% and 58% of the total and blood culture isolates of S. aureus in Hong Kong public hospitals are MRSA, respectively [4]. MRSA is getting more resistant to antibiotics that are currently used, and even a glycopeptides resistant strain, Vancomycin-resistant S. aureus (VRSA) has also been reported [5]. Similarly, MTB, the causative agent of tuberculosis, is still a major potential threat. Globally, an estimation of 9.6 million people had suffered from TB, and among these, about 16%, or 1.5 million, were killed [6]. It was estimated that 3.3% of new TB cases are multidrug-resistant tuberculosis (MDR-TB), and 20% of previously treated cases are MDR-TB, which are significant numbers [6]. MDR-TB is extremely difficult to cure, with a treatment success rate of merely 50% [6]. Furthermore, an even more resistant form of TB, totally drug-resistant TB (TDR-TB) has been described [7]. With resistance to all tested drugs, TDR-TB might be untreatable by any currently available drugs [8]. Therefore, there is a definite need to develop new approaches and druggable targets for combating drug-resistant pathogens.
A novel approach to solve the problem of drug-resistance is to “disarm” the microbes [9]. Instead of being bactericidal or bacteriostatic, the new drugs would inhibit virulence factors of pathogens, and control the infection only by lowering the virulence of the pathogens. Virulence of a pathogen refers to its ability to cause diseases and is contributed by a variety of factors that lead to damage in the host [10]. These factors, for example, include molecules that assist in host-recognition, toxins that damage host tissue, adhesins that assist adhesion, and secretion system-related molecules that assist the release of other factors [10,11]. S. aureus pathogenesis depends on a specialized protein secretion system (type VII-like Ess) that delivers a range of virulence factors to assist infectivity by establishment of abscess lesions and suppression of host immune responses [12,13]. EsxA is a confirmed excretory virulence factor of S. aureus [14]. EsxA is reported to be an immune evasion gene but how it can achieve this function is still unclear [15]. EsxA is small alpha-helical polypeptides belonging to a family of WXG100 motif proteins [16], which have a size of approximately 100 amino acids containing a helical structure and a conserved TrpXaa-Gly (WXG) motif [17]. ESAT-6 (homolog of S. aureus EsxA) and CFP-10 (homolog of S. aureus EsxB), secreted by Mycobacterium tuberculosis (MTB), are the founding members of the WXG100 motif family. ESAT-6 and CFP-10 are early secreted virulence factors by Mycobacterium tuberculosis, the causative agent of tuberculosis (TB) [18,19]. These virulence factors are crucial for the replication of MTB in macrophages and presumably also for the pathogen’s ability to suppress innate and adaptive immune responses [19- 21]. Mutants that fail to secrete EsxA or EsxB display defects in the pathogenesis of S. aureus murine abscesses, suggesting that these specialized secreted WXG100 motif proteins may be a general strategy of human bacterial pathogenesis [14]. Therefore, EsxA is a promising drug target for combating MRSA and drugs targeting EsxA may have “add-on” implications in fighting against other drug resistant pathogens such as multidrug-resistant strains of MTB. In the present study we have streamlined a platform to perform in silico structure-based screening on a large ligand library containing 6.8 million compounds to identify lead compounds targeting the virulence factor EsxA of S. aureus.
Materials and Methods
In Silico Structure-Based Screening on EsxA and Clustering Analysis
In silico structure-based virtual screening on S. aureus EsxA (PDB ID 2VS0) against a library of Lead-Like subset of all purchasable compounds in the ZINC database (https://zinc.docking.org/) containing 6,746,897 compounds and a focused library of 50,240 bioactive compounds (ChemBridge Corporation) were performed on the High Performance Computing (HPC) Linux cluster at the University of Hong Kong. Autodock Vina (http://vina.scripps.edu/) [22] was used to identify optimal binding poses for the docking of each ligand to the S. aureus EsxA. The ligands were allowed to have all torsions rotatable except the rings during docking simulations. The docking search grid dimensions were 80Å × 80Å × 80Å along the x-, y- and z-axes to cover the whole S. aureus EsxA protein. The center of the docking ligand was initially positioned at the grid center. Grid searching was performed using the Lamarckian genetic algorithm with other parameters set at default values. A suite of in-house scripts has been developed to perform tasks of preparing each ligand for docking, the parallel submission of molecular docking jobs to the computing queues in HPC cluster to maximize throughput as well as sorting and ranking the results according to the docking scores reported. The web-based tool, Automatic analysis of poses using self-organizing maps (AuPosSOM) (https:// www.biomedicale.univ-paris5.fr/aupossom), was used to perform further clustering analysis on the docking results. All calculations were performed using default parameters. The best leaves of the all contacts and hydrogen-bond contacts main trees were further undergone subtree analysis to identify common and successful binding modes. The best 100 consensus scoring compounds were selected to create a list of putative hit compounds for secondary screen and validation by NMR titration assay.
Expression and purification of EsxA
The plasmid of pET2wb-esxA was transformed and expressed in Escherichia coli strain BL21 (DE3). Recombinant protein expressions were started by picking a colony from a plate of transformed cell, to 10 mL of sterilized LB medium with a concentration of 100 µg/mL ampicillin (LB/Amp+) for growth at 37 °C for 16-18 hours. This starting culture was then transferred to 1L LB/Amp+ medium for growth at 37 °C with agitation of 220 RPM till the mid-log phase (OD600 = 0.6-0.8). The expression of His-tagged EsxA was triggered by addition of 0.1mM IPTG into the cell culture and hereby inoculated for 4-5 hours at 37 °C. The induced cells were harvested by centrifugation at 4000 RPM for 30 minutes at 4 °C. The cell pellet was resuspended and was lysed by sonication. This lysate was then subjected to centrifugation at 18000 RPM at 4 °C for 30 minutes. Only the supernatant of cell lysate was collected for subsequent purification. The recombinant protein was first purified with a Ni2+ column according to the manufacturer’s protocols. The protein was further purified by size exclusion chromatography on a Superdex HiLoad 16/60 S75 prep grade column. For unlabelled EsxA, normal LB medium were utilized as the culturing broth. For 15N-labelled protein samples, M9 medium supplemented with uniformly labelled 15N-NH4 Cl was used to limit the nitrogen sources in the culture medium.
NMR Titration Experiments of EsxA with Hit Compounds
To experimentally validate the docking results, the proteinligand interactions between EsxA and each of the top 100 scoring putative hit compounds were validated by NMR chemical shift perturbation titration method. 1H-15N HSQC spectrum of a 15N-labelled 0.3 mM EsxA sample was monitored for either crosspeak movements or intensity variations with the titration of an unlabelled hit compound into the EsxA sample.
Susceptibility Tests on List of EsxA Hit Compounds
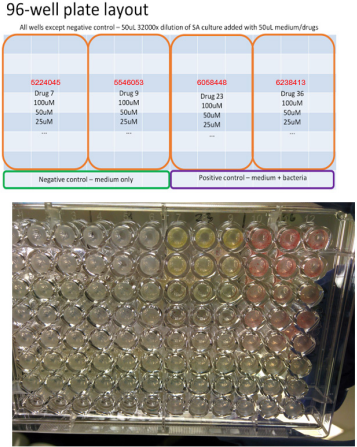
MRSA strain Mu3 was tested for susceptibility of those hit compounds with validated interaction with EsxA by NMR titration. Briefly, colonies of S. aureus strains were picked and streaked on BHI agar plates and incubated at 37 °C overnight for 16h. Afterwards, single colonies of the bacteria were picked and put into culture tubes together with a 2mL culture solution, which is Mueller Hinton broth with 0.1g/mL CaCl2 and MgCl2 added. The bacteria were then cultured at 37 °C in a shaker for 6h. The 96-well plates for the broth microdilution test were prepared according to Supplementary Figure S2. Each experimental well contains 50 uL of 32000-fold dilution of the prepared bacteria culture, 49 uL of culture solution, and 1 uL of test compound dissolved in DMSO in various concentrations. 100 uL of culture solution serves as negative control, while 50 uL of MHB and 50 uL of 32000-fold dilution bacteria culture serves as positive control. Serial 2-fold dilution of the test compounds were performed in a separate 96-well plate, using DMSO as solvent. 20 uL of each compound at 10 mM were prepared as the starting concentration, then 2-fold serial dilution was performed using DMSO up to 64-fold dilution, which are then added separately to the experimental wells. The plates were cultured at 37 °C for 16h overnight. The wells were then observed visually to determine any bacterial growth, as indicated by cloudiness of the solution or growth of cells at the bottom of the wells.
Supplementary Figure S2: Susceptibility test of hit compounds (ID 5224045, 5546503, 6058448, 6238413) against MU3.
Virulence Assay on Validated List of EsxA Hit Compounds
Given that EsxA is a virulence factor of S. aureus, the effect of the validated hit compounds on the virulence of MRSA was tested by observing the change in expression of virulence genes. Two types of transformed bacteria were prepared, US300-pGLspa2 and US300- pGLhla. They are the MRSA strains US300, transformed by luciferase plasmids encoding Staphylococcal protein A promoter (pGLspa2) and alpha-toxin promoter (pGLhla), respectively. As the plasmids also encode a chloramphenicol resistance gene, the bacteria culture stock from -80 °C freezer were streaked onto BHI agar containing 10 ug/mL chloramphenicol incubated at 37 °C for 16h for selection. 5 isolated colonies from each type of bacteria were then picked and cultured in 2mL BHI with 10ug/mL chloramphenicol for 16- 18h in 37 °C shaker. The lux was then measured for each type of the cultured bacteria, and the tube giving highest lux was taken to culture for a further 6h at 37 °C shaker. Then, disk diffusion test sterile paper disks were prepared and aligned on plates spread with the test bacteria. Lastly, after incubating the plates at 37 °C for 16h, the results were visualized using PerkinElmer IVIS Spectrum In Vivo Imaging System.
Results and Discussion
In Silico Structure-Based Screening on EsxA and Clustering Analysis
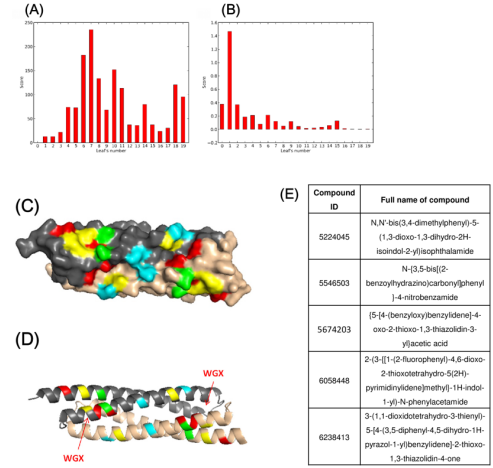
In the present study, a dedicated research platform with the capacity for performing in silico structure-based screening of large ligand library on a target virulence factor protein was setup. Autodock Vina dockings were completed on all 6.8 million ligands in the library against the X-ray structure of EsxA (PDB 2VS0). Docking score sorting and clustering analysis by AuPosSOM of the best-hit ligands according to their structural features and poses were performed in order to draw up the list of best docking scoring ligands for secondary screening and validation. Figure 1A-1D showed the AuPosSOM scoring plots and the graphical representation of the major predicted contacts on EsxA dimer structure.
NMR Titration Experiments of EsxA with Hit Compounds
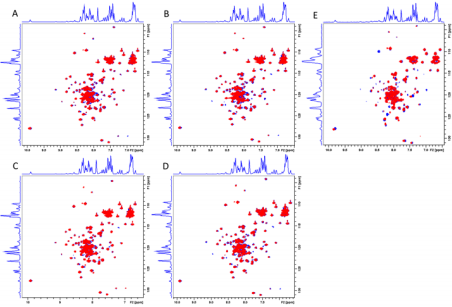
Out of the 100 top hit compounds, 5 compounds with IDs and IUPAC names shown in Figure 1E could produce significant cross-peak movements or intensity variations upon titration into 15N-labelled EsxA in secondary experiment screen by NMR titration assay (Supplementary Figure S1).
Figure 1: Clustering analysis of docking results and list of validated hit compounds.
A) AuPosSOM scoring plot using all contacts for analysis. Each leaf number corresponds to a particular cluster of compounds with similar binding modes. A high positive score indicate high chance of finding active compounds inside the leaf;
B) AuPosSOM scoring plot using hydrogen bonding contacts for analysis.
C) Graphical representation of major predicted contacts on surface plot of EsxA dimer structure (PDB: 2VS0).
D) Graphical representation of major predicted contacts in ribbon plot. Red: Major contacts exclusive to leaf 6 of all contacts main tree; Green: Major contacts exclusive to leaf 7 of all contacts main tree; Yellow: Major contacts discovered in both highest scoring leaf 6 and 7 of all contacts main tree; Cyan: Major contacts that are exclusively found in hydrogen bond analysis.
E) List of hit compounds that displayed binding effect in secondary experiment screen by NMR titration assay with EsxA.
Supplementary Figure S1: Overlaying of 1H-15N HSQC spectra of hit compounds that show interaction with EsxA. Blue: 2D spectrum of apo-EsxA sample; Red: 2D spectrum with addition of hit compound.
A) Hit compound ID 5224045
B) Hit compound ID 5546503
C) Hit compound ID 6058448
D) Hit compound ID 6238413
E) Hit compound ID 5674203.
Susceptibility Tests on List of EsxA Hit Compounds
The broth microdilution test showed that hit compound 6058448 showed MIC at 25 uM while hit compounds 5224045, 5546503, and 6238413 have MIC >100 uM (Supplementary Figure S2).
Virulence Inhibition Assay on List of EsxA Hit Compounds
Only 1 hit compound 5674203 showed significant antivirulence effects by inhibiting the expression of both protein A and alpha-toxin virulence factors while the other 4 hit compounds 5224045, 5546503, 6058448, and 6238413 did not show any inhibitory effects (Figure 2).
Figure 2: Virulence inhibition assay on hit compounds against MRSA strain US300. Middle circle is the positions of the hit compounds (ID 5224045, 5546503, 5674203, 6058448, 6238413) in each of the assay dish. Top panel is the inhibition assays against US300-pGLspa2 transformed by luciferase plasmids encoding protein A promoter at 5mM and 500uM. Bottom panel is the inhibition assays against US300-pGLhla transformed by luciferase plasmids encoding alpha-toxin promoter at 5mM and 500uM.
Conclusion
The local and global spread of MRSA is of great concern because it is quickly acquiring resistance to all clinical antibacterial agents. Therefore, there is an urgent need to develop new approaches and new druggable targets to combat MRSA with lower chances of drug resistance development. In this study, we have identified several hit compounds against MRSA through in silico structure based screening against the novel target EsxA of S. aureus. These hit compounds will likely act against MRSA through a mechanism different from the existing antibiotics because EsxA is a virulence factor of S. aureus. For example, hit compound 5674203 showed no antibacterial effect but significant anti-virulence effects to enhance host cell protection. Anti-virulence drugs against S. aureus infection with diminished virulence will cause less or even no damage to the host cells and tissues and will not subject to natural selection pressure so that the anti-virulence drug will less likely cause drug resistance. Through the hit-to-lead process, these hit compounds can be further modified and improved to attain higher potency and better pharmacokinetics properties. The improved compounds could be the candidates for new drug development against MRSA. Through the same approach, we have also obtained promising hit compounds against the ESAT-6 and CFP-10 virulence factors of MTB, which can effectively suppress the intracellular survival of MTB model bacteria Mycobacterium marinum in macrophages without directly killing the bacteria (to be published in a forthcoming manuscript). Therefore, the present platform for in silico structurebased screening of large ligand library can be readily applied to other important drug targets or virulence factors with known highresolution structures.
For more Articles on : https://biomedres01.blogspot.com/



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.