High Resolution Melting Curve for the Rapid and Efficient Detection of SARS-Cov-2 Gene Variation in the Greek Population
Introduction
As of May 6, 2021, approximately 153 million cases of Coronavirus
disease 19 (COVID-19) have been reported worldwide [1]. Due to
the rapid spread of the causative SARS-CoV-2 corona virus, the
development of a quick and accurate detection assay is considered
vital aiming to control the possible sources of infection, in order
to design effective measures to prevent further transmission.
Routine laboratory confirmation of SARS-CoV-2 infection is based
on nucleic acid amplification tests (NAAT), such as the real-time
reverse transcriptase-PCR (RT-PCR), which is adopted as a simple
qualitative assay that combines a relatively high sensitivity [2]
with high specificity for the detection of the virus [3,4]. The most
commonly used targets for primer/probe development derive
from the conserved viral genome, including the ORF1ab, RNA
dependent RNA polymerase (RdRp), nucleopcapsid (N), envelope
(E) and spike protein (S) genes [5-7]. Although SARS-CoV-2 quasispecies
variants can be most accurately identified through whole
genome sequencing or alternatively by the means of Sanger or next
generation sequencing amplicon-based sequencing of selected
parts of the viral genome [1], these methods are regarded both as
time consuming and expensive.
Thus, a simple and more rapid screening approach is needed to be developed, for the key SARS-CoV-2 mutations that define variant strains. High Resolution Melting (HRM) is a novel, homogeneous, close-tube, post-PCR method, enabling genomic researchers to analyze genetic variations either as single nucleotide polymorphisms (SNPs), point mutations, or methylation degree in PCR amplicons. This assay prevails the power of a classical melting curve analysis by enabling a detailed study in the thermal denaturation of a double-stranded DNA, providing us finally sufficient information. In this study we tested the possibility that a Real-Time PCR combined with HRM assay will serve as useful and efficient diagnostic technique for the detection of SARS-CoV -2 variables in 4 viral genome locations, namely RdRp, N1, E and S2 genes. The High-Resolution Melting Curve was performed in a post RT-PCR assay, and amplified a fragment of these genes, in order the generic diversity of the SARS-CoV -2 can appropriately be examined. Our hypothesis was that this method can be adopted as a valuable tool for the rapid screening of large numbers of patient samples for the tested variants, providing an early warning for the emergence and spread of these strains of concern.
Materials and Methods
Clinical Samples
A total of 620 clinical specimens collected in the municipal area of West Attica and sampling was accomplished to our premises with all the required precautions. All volunteers were suspicious COVID-19 cases, according to World Health Organization criteria (World Health Organization, 2021c). We only collected nasopharyngeal swabs, which subsequently were placed in 2 ml of transport medium with neutralizing agent. We used Disposable Virus Sampling Tube (Zybio; Inc; China) which adopts efficient virus inactivation technology and special flocked swab. It can be used for the collection and storage of clinical novel coronavirus, influenza, avian influenza (such as H7N9), hand-foot-mouth virus, measles and other virus specimens, as well as for chlamydia, mycoplasma, and ureaplasma. Specimen processing was performed in a class II biological safety cabinet using biosafety level three (BSL3) work practices.
RNA Extraction
Nucleic acids were recovered from clinical specimens using an automatic extractor (MagDEA DNA / RNA 200 virus), The RNA samples were separated in two aliquots.
qRT PCR
The first aliquot was used for the qRT-PCR using the Mutaplex SARS-CoV-2 commercial kit (Immundiagnostik AG). Specific primers were used for highly conserved regions and double-labeled probes to enhance and differentiate RNA SARS-CoV-2 and other beta Coronaviridae such as MERS. Detection of SARS-CoV-2 was visualized at the FAM / GREEN channel. Beta-coronaviruses (SARSCoV- 1 and SARS-CoV-2) are detected at Cy5 / RED channel. Internal Process Control (IPC), which was added during RNA extraction, was detected in the same reaction at HEX/ YELLOW. Detection of RNA Polymerase (human gene) allows RT-PCR detection of inhibitors confirming in addition viral RNA was isolated from specimen.
cDNA Synthesis
The second aliquot was used for the cDNA synthesis following a qRT-PCR with a post High Resolution Melting Curve. cDNA was synthesized using Luna Script® RT SuperMix Kit (New England Biolabs). A 5 μL aliquot of purified RNA was added to 4 μL of the Luna Script® RT SuperMix Kit. The reaction was performed in a total volume of 20 μl.
qRT-PCR and HRM Assay
5 μL cDNA was added to 20 μL of six different reaction mixtures containing 500 nM each primer. The primer sequences used for RNAdependent RNA polymerase gene detection were FW 5’AGA-ATA-GAGCTC- GCA-CCG-TA3’and REV 5’ CTC-CTC-TAG-TGG-CGG-CTA-TT-3’ giving an amplified product of 101bp. The primer sequences used for E gene detection were FW 5’TTCGGAAGAGACAGGTACGTTA-3’ REV 5’AGCAGTACGCACACAATCG-3’ giving an amplified product of 116bp. The primer sequences used for N1 gene detection were FW 5’CAATGCTGCAATCGTGCTAC-3’ and REV 5’GTTGCGACTACGTGATGAGG-3’ giving an amplified product of 117bp. The primer sequences used S2 gene detection were FW 5’GCTGGTGCTGCAGCTTATTA-3’ and REV 5’AGGGTCAAGTGCACAGTCTAA-3’ giving an amplified product of 107bp. 25-μL reaction was setup that contained 5 μL of cDNA, 12.5 μL 12.5μL Melt Doctor master mix, which includes HRM dye (Melt Doctor Applied Biosystem).
PCR Cycling for HRM Curve Acquisition was Run Under the Following Conditions
One cycle at 95°C for 10 min; 40 cycles at 95°C for 15 s, 60°C for 40 s, and 72°C for 30 s; Then the fragment was melted by raising the temperature from 60°C to 95°C, with an increment of 0.11°C/s, in order to obtain information on melting profiles. Melting-curve analysis was performed using the HRM software of Applied Biosystems This software analyzes the HRM curve data to identify changes in the shape of the curve that indicate sequence polymorphisms.
PCR Test for cDNA Quality
As an optional step, a cDNA quality test was performed after cDNA synthesis to verify the appropriate synthesis of the cDNA from each sample. GAPDH genes of Homo sapiens with primer set FORW 5’ CAA-TGA-CCC-CTT-CAT-TGA-CC.3’ and REV 5’ TTG-ATTTTG- GAG-GGA-TCT-CG was used for the human IPC. In this PCR protocol, 5 μL of cDNA was used with the following PCR cycling conditions: 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 60 °C for 40 s, and 72 °C for 1 min, with the final elongation step at 72 °C for 5 min. Each primer was used at a concentration of 500 nM in 2× PCR premix reagent (Promega Hot Start Green Master Mix). The amplicons were subjected to electrophoresis in a 2% agarose gel at 130 V for 20 min and visualized under UV light giving an amplicon with 159bp.
Standard Curve and Limit of Detection (LOD)
The real-time PCR with Melt Doctor standard curve was generated by serial 10-fold dilutions of synthetic positive controls in RdRp, E, N1 and S2 genes with known copy numbers (10.000, 1.000, 100, 10 and 1 copies/μL). These dilutions were tested using 10 replicates and they were used as quantification standards to construct the standard curve by plotting the copy number against the corresponding threshold cycle values (Ct). To verify the specificity of the reaction, the melting curve analysis and electrophoresis on agarose gel were carried out for the products of the real-time PCR reaction. Five microliters of the amplicons were electrophoresed in 2% (w/v) agarose gel stained with ethidium bromide in 1x (TBE) buffer and visualized by ultravioltet (UV) light in order to check All specimens were aliquoted at reception and those not used in the assays were stored at -800C for later confirmation of PCR results. Positive results were considered valid when the PCR results matched in two different aliquots and analyzed with the commercial kit and the real-time PCR with HRM assay.
Biomedical Ethics Issues
The collection of clinical data from volunteers, were correlated with the laboratory research results and were conducted in such a way as to fully guarantee the patients’ anonymity and personal data confidentiality.
Statistical Analysis
Statistical analysis. Standard statistical analyses (average, standard deviation, correlation coefficient) and graphing were performed using Microsoft excel (ver 2102) for Windows.
Results
The linearity and efficiency of the real-time PCR were determined by generating a standard curve in which serial 10-fold dilutions of positive control were tested. The standard curve was generated by plotting the real-time PCR threshold cycle numbers (Ct) of each dilution against the known copy numbers of positive control. The resulting slope showed a linear relationship over 5 orders of magnitude, ranging from 10.000 to 1 copies/μL with a correlation coefficient R2>0.99. The detection rate was 100 % for up to 2,5 copies/μL having 10/10 replicates positive for E and N genes and 5 copies/μL having 10/10 replicates positive for S and RdRp genes. Strong linear correlations (r2 ≥0.99) were obtained between CT values and transcript quantity. Assay reproducibility and repeatability was tested by using replicate 10-fold serial dilutions of the RNA transcripts evaluated for each dilution point in triplicate on three different days. At the lower copy detection limit for SARS-CoV-2 and assay reproducibility exceeded 95%. Over the linear range of the assay, the coefficient of variation of the mean CT values within and between runs was 0.46%–2.54% and 0.64%– 2.39%, respectively.
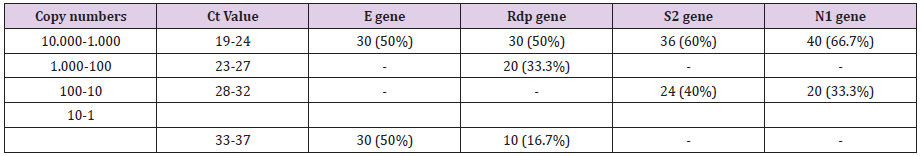
The methods have thus been shown to be highly capable of detecting the novel SARS-CoV-2 with 100 % specificity. The specificity (100%) of the reactions was confirmed by a melting temperature for positive control dilutions, indicating the formation of a single PCR product with no artefacts, such as nonspecific amplification products or primer dimers (results not shown). Furthermore, amplification products were also checked on agarose gel stained with ethidium bromide in standard TBE buffer and clear and well-defined specific bands with the expected sizes for all replicates of positive control dilutions. All 620 RNA samples tested, were positive for the human gene (GADPH) which was included as internal control to evaluate the quality of clinical specimens (nasopharyngeal swabs) and nucleic acid extraction. From the 620 RNA specimens, 60 were tested positive for SARS-CoV-2 indicating a prevalence of 9,7% by both methods. The Ct Value ranged from 19 to 36 that corresponded to 10.000- 1 copy numbers. The qRT-PCR and the real-time PCR with HRM assay displayed 100% sensitivity.
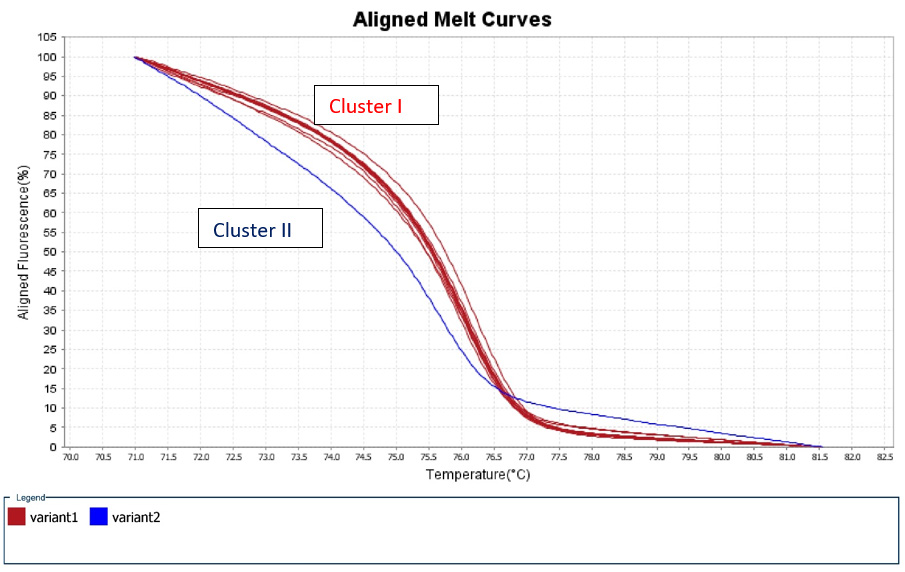
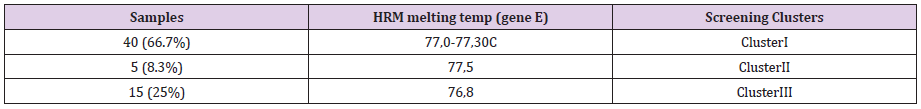
A 50% of the specimens tested comprised 10.000 to 1.000 copy numbers of E and RdRp amplicons. Sixty and 66,7% of the specimens tested, contracted 10.000 to 1000 copy numbers of the S2 and N1 genes, respectively. A range of 1000 to 100 copy numbers was also identified in 33.3% of the specimens for the RdRp gene. A range of 100 to 10 copy numbers was further identified in 40 and 33.3% of the specimens for the S2 and N1 genes, respectively. Finally, a range of 10 to 1 copy numbers was identified in 50 and 16.7% of the specimens of the E and RdRp genes. All the sixty positive for the virus-RNA samples were synthesized to cDNA following a qRTPCR with a post High-Resolution Melting Curve. The HRM diversity assay was used to analyze the regions of the four genes. As shown in Tables 1-4 and Figure 1, all SARS-CoV-2 positive samples produced measurable Tm values. The E gene demonstrated the highest variation with three clusters, with 66.7% belonging to cluster I. N1 gene amplicons identified two clusters with 50% of the specimens in each cluster. The S2 gene did not show any variation, forming only one cluster. Similar results were shown for RdRp gene, where 95% of amplicon belonged to one cluster (Table 5).
Figure 1: High Resolution Melt Curve This is an aligned melt curve. The plot demonstrates the sharp decrease in fluorescence when the double-stranded DNA melts into its single-stranded form. Diversity of positive specimens of gene Rdp.
Discussion
SARS-CoV-2 is a highly transmissible and pathogenic coronavirus that emerged in late 2019 and has caused a pandemic of acute respiratory disease, named Covid-19, which threatens human health and public safety. Little is known about the genetics of this virus but the health communities have to respond to any new viral genetic variants, pursuing immediate countermeasures. However, it is inevitable that in a limited number of individuals a new virus population during transmission will be established. Thus, remains crucial the concerning variants to be early detected by the means of rapid and effective assays and therefore we need ascertained genomic epidemiology projects for large scale monitoring of the SARS-CoV-2 evolution. High Resolution Melting Analysis is a molecular technique based on RT-PCR that has been largely used to rapidly detect other pathogen strains, resistant to treatment. We developed the first homemade RT PCR combined with high resolution melting curve for the detection and screening of Sars-CoV-2 variants.
Although detection of the viral nucleic acid using an RT-PCR assay has become a standard and formative assessment for the diagnosis of COVID-19 [8,9] the main concern in RT PCR are the false negative results usually attributed to the low quality of the specimen and to the inappropriate sample handling. In order to be ensured that our samples had a suitable genetic material for RTPCR and adequate portion for HRM would be then extracted, we used Internal Positive Control primer sets of GADPH genes of Homo Sapiens [10,11]. All of the 620 samples studied were positive for GADPH gene, confirming therefore the quality of the extracted RNA and the converted cDNA portions. The quantification of SARS-CoV-2 RNA in clinical specimens by reporting Ct values and copy numbers of RT-PCR is generally limited [12], but this is due to the current needs of diagnosis. Efficacy of RT-PCR in the diagnosis of SARSCoV- 2 infection is greatly dependent on the pre-analytical phase, including the patient selection and material collection.
Even more the extraction method of RNA and the performance of RT-PCR test kit [12] interfere significantly with the results. Although Ct values are affected by a number of factors, they may be still able to provide important clinical data for decision making by providing an indication of viral load [13]. A positive correlation has been demonstrated between the viral load and either the severity of COVID-19, or the intensity of hypoxemia, the risk of death, or with various other hematological, biochemical, and inflammatory alterations [14,15]. As we were not aware of the complete medical history for the majority of the volunteers, the detected low Ct numbers in the 50% of the examined specimens. This probably corresponds to a high viral load of the sample which was taken 3-6 days following the symptoms onset from individuals residing in an area with high COVID-19 prevalence. Most of our study participants did not suffer from any underlying diseases and they received home-based treatment, while none them was hospitalized for any reason. In specimens that were taken 15 days later, it was shown to have higher or negative Ct values, in comparison to samples taken in the beginning of the infection (data non shown), indicating somehow a recovery status.
Although reverse transcription PCR remains the most sensitive and accurate method for the detection of the new coronavirus, the use and the sufficient supply of commercial kits for this purpose may not considered as a cost-effective solution [16]. Currently RT PCR using different sets of primers and dyers, is applied for the detection of variants [13,11,12]. However, HRM assay seems to be a promising tool, when it is combined with RT-PCR, even without any other previous testing. Even more HRM technique can be noted as a rapid, low-cost, and, high-throughput method for quantifying genetic diversity. Using thermal denaturation of double-stranded DNA, the technique extracts significant details and is capable of finding SNPs. With this method we are able to identify smaller differences in PCR amplicons, down to the single base level and therefore this method is accepted as an ideal procedure for single nucleotide polymorphism genotyping, species identification, sequence matching and mutation scanning, without the need for any further separation and additional processing following a PCR. Several HRM assays have already been successfully conducted in the past and have been applied for the detection and genotyping of viruses such as HIV[17], astroviruses [18], polyomaviruses [19], noroviruses [20] and influenza A viruses [21].
To our knowledge, no study has been conducted as yet, using high-resolution melting analysis (HRM) technique, for the rapid detection of variations within S, N, E and RdRp genes of SARS-CoV-2. In our study the highest variation with three clusters was identified in the gene E which encodes the Envelope protein E proteins, that help in the assembly and release of the virions [22]. The E protein plays important roles in viral morphogenesis, replication, and pathogenesis [23] and is conserved in coronaviruses [24]. Among the structural proteins of the SARS-CoV-2, E protein is considered as a potential drug target. However, according to the GISAID database (as of 25th May 2020), more than 40 amino acid mutations of the E gene were found from 4085 SARS-CoV-2 genomes. The E gene of SARS-CoV-2 seemed to have a high mutation rate and thus it would be difficult to synthesize an effective antiviral molecule aiming to E gene expression which may be carrying a diverse population of different strains.
Nucleocapsid proteins (N) play an important role in the packaging of viral RNA and mediate viral assembly by interacting with the viral genome and M protein, which are helpful in the augmentation of viral RNA transcription and replication [25]. Based on the high sequence similarity of N protein within the coronavirus family, it may be suggested that antibodies against the N protein of SARS-CoV would likely recognize the N protein of SARS-CoV-2. Although our results indicate variations divided in 2 distinct clusters, N gene show highly conserved regions and therefore N proteins are also considered as potential drug targets. Nevertheless, the clinical relevance of N2 negativity is considered to relate with asymptomatic or subclinical disease course as was the case of our volunteers [26]. S gene encodes the spike (S) protein which interacts with angiotensin-convertin enzyme 2 (ACE2). The inhibition of this association is a possible target for the development of novel therapeutic approaches [27]. The S2 subunit mediates the viral cell membrane fusion and the detected gene stability in our study is encouraging the idea that inhibiting the gene expression, would serve as a potent therapeutic intervention with constant effect for the prevention of disease transmissibility and pathogenesis.
The lack of any S2 variability seen in our study is providing confidence that we will not face a potentially compromised vaccine effectiveness in Greece for the near future, since S protein serve as the major viral antigen for the current vaccines. RNA-dependent RNA polymerase (RdRp), is considered a promising but challenging drug target for inhibiting replication and hence, the growth of various RNA-viruses. The widely used antiviral drug Remdesivir has an anti-RdRp activity and various under development other potent prospective drug candidates against the SARS-CoV-2 are targeting RdRp proteins. The absence of any variations in our results in regard of RdRp gene empower the steady effect of this therapeutic approach in mitigating the disastrous global effects of the COVID-19 pandemic. The emergence of SARS-CoV-2 variants which may be proved potential for increased transmission, disease severity, and resistance to vaccine induced immunity is of grave concern. A simple screening assay to monitor the emergence and spread of these variants may be helpful for epidemiological studies. With our study it was demonstrated that our assay using the highthroughput PCR assay platform is a simple, rapid, and sensitive and specific tool for detecting variant-identifying mutations.
Our HRM assay may reinforce the further investigation of the novel coronavirus diversity and the detection of newly emerging virus variants. High-Resolution Melting Assay will also possibly lead to a reduction in the need of sequencing techniques by the exclusion of the samples with the same HRM curves. Therefore, it could be then adopted as an inexpensive molecular tool for public health screening studies in order to pursue SARS-Cov-2 variants. Probably the most important limitation of the method is that it may not reveal all the sequence variations in a cDNA fragment. The limitations regarding the detection method of our home set up RTPCR was regarded similar to the commercial kit used in primary diagnosis of SARS-CoV-2 2 infection in the routine laboratory. Another limitation of the study is the rather small sample study, but the need for the prompt circulation of all relevant information regarding the SARS-CoV-2, convinced us to submit our preliminary results. Our data may then facilitate the design of a universal admission screening course.
Conclusion
A home-made RT-PCR, determining separately the expression
of four gene targets, is useful for the current needs of SARS-CοV-2
laboratory diagnosis. Post RT-PCR, High Resolution Melting curves
(HRM) could be considered as a rapid screening method for the
identification of variants and strains of concern. Improvements of
this method and further research are required in order to monitor
effectively the virus evolution and the corresponding host immunity.
For more Articles on : https://biomedres01.blogspot.com/




No comments:
Post a Comment
Note: Only a member of this blog may post a comment.