Possible Use of CRISPR-Cas13 Technology in the Pathogenesis of the SARS-CoV-2 Virus
Introduction
In order to start thinking about this alternative, it is important
to know the mechanism of action of viruses in order to identify
the attack sites of the technique. It is important to know that the
infection begins in the host cell, when the virus binds to a receptor
on the cell surface. The SARS-CoV-2 virus is bound by its protein
(S) and the receptor for angiotensin converting enzyme 2 (ACE2).
This binding directs the north of the virus in the target cell and its
specificity [1]. ACE2 contributes to the regulation of blood pressure
by converting angiotensin I into angiotensin [2]. The ACE2 receptor
is expressed in many tissues such as the lower respiratory tract,
heart, kidney, stomach, bladder, esophagus, and intestine [2]. In
the lung, it is expressed in the alveoli [3]; and in the oral cavity, in
epithelial cells of the tongue [4]. The SARS-CoV-2 protein (S) has
two subunits (S1 and S2). The S1 subunit is the one that interacts
and binds to the ACE2 receptor through the receptor-binding
domain (RBD), while the S2 subunit determines the fusion of the
virus membrane with that of the host cell [5].
The virus completes its entry into the cell when the S protein
is cut by the protease enzyme (TMPRRS2). This generates the
separation of the RBD union of the S1 subunit with the ACE2
receptor and its fusion with the membranes, and in this way the
virus is left with the door of entry open to the cell by endocytosis
[6]. Once inside the cell, the translation of the viral genome and the
transcription of the SARS-CoV-2 proteins begin. When it enters the
cytoplasm, the nucleocapsid of the virus is released and the viral
genomic RNA is released. This RNA acts as an mRNA so the viral replicase gene can be directly transcribed [1,7]. To generate the
necessary proteins of the replicase transcriptase complex (RTC),
which is assembled in the endoplasmic reticulum (ER) ([1,5,7-11].
Finally, the complex (RTC) replicates and synthesizes a set of sub
genomic mRNA (sgRNA) [8-11] that code for the elaboration of the
main structural proteins (S), (M), (E), (N) and accessory proteins
[1-5].
So, this is how RNA replication, Protein Assembly and SARSCoV-
2 Egress from the Host Cell are built. In SARS-CoV-2 replication,
single-stranded RNA (+ ssRNA) serves as a template to initially
synthesize a copy of single-stranded RNA (-ssRNA) [12]. And from
this copy of -ssRNA, polyproteins will be produced, which will make
up the RTC complex [1,8]. The RTC complex again creates a copy
of the original virus genome + ssRNA from the -ssRNA template.
This associates with the protein (N) forming the nucleocapsid. The
structural proteins (S), (M) and (E); and accessory proteins from
the endoplasmic reticulum (ER) and then, in the Golgi complex, are
assembled together with the nucleocapsid to produce new viral
particles, which are produced towards the plasma membrane for
the release of the virus [5]. According to this final mechanism, it is
where the grouped and regularly spaced short palindromic repeats
(CRISPR) emerge as a therapeutic alternative, together with the
endonuclease Cas, form the CRISPR / Cas complex.
This system was discovered as an immune defense mechanism
present in bacteria and archaea, which incorporate DNA from
pathogens, such as bacteriophages, between repeated palindromic
sequences and subsequently generate an RNA called “crRNA” when
transcribed. Due to its activity as an endonuclease and recognition
capacity in specific sequences, the CRISPR / Cas system has been
exploited in genetic engineering to activate genes, repress them,
induce point mutations and change sequences through homologous
recombination. CRISPR has also been used to generate murine
models of human diseases and to evaluate cell physiology through
the simultaneous activation or repression of various genes [13,14].
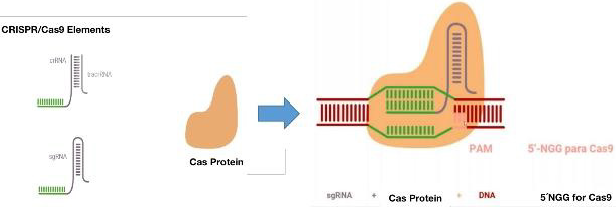
This mechanism is made up of two factors, an endonuclease and
a complementary recognition sequence. Thus: an RNA from the
CRISPR sequence, called “crRNA”, and the Cas endonuclease. The
crRNA is in charge of directing Cas towards its complementary
sequence, where Cas makes the cut.
The CRISPR sequence is composed of a leader or promoter
and different spacer sequences of more or less than 40 nucleotides
on average and side and side repeated sequences called palindros
with an extension of approximately 32 nucleotides Figure 1 [15].
When this mechanism enters into action in the cytoplasm, the
cell recognizes a sequence known as a motif adjacent to the PAM
protospacer and incorporates the nucleotides adjacent to the PAM.
Figure 1. Subsequently, the crRNA is transcribed. The Cas then
associates with the mature crRNA and forms the CRISPR / Cas
complex. The crRNA will be the one who guides the complex towards
its target through the recognition of the complementary sequence.
The cuts produced by the explained mechanism are repaired
by non-homologous end joining (NHEJ) and homology directed
repair (HDR). As NHEJ can cause unwanted effects of unexpected
mutations, it is preferred to choose HDR for this method. This
CRISPR / Cas complex is a good alternative for targeted genomic
editing.
The system has made it possible to insert, eliminate or
generate mutations in the sequences, which leads it to target a
specific sequence and induce a cut in both DNA strands [13,14].
However, this system would not be very relevant for SarsCov2,
since it is an RNA virus, but within the range of possibilities and
versatility of the CRISPR system, a subsystem of this complex,
known as CRISPR-Cas13, has been identified. CRISPR-Cas13 are
widely distributed among Leptotrichia species. Estimating the
occurrence, composition, and diversity of CRISPR-Cas13 systems in
the genus Leptotrichia is particularly challenging because complete
information on the entire genome is limited [16]. Cas13 is a crRNAguided
RNA-targeted effector, which has two distinct RNase sites [17]. Not only does it highlight its unique RNA-directed ssRNA lysis
activity, but Cas13a becomes a promiscuous RNase that can nonsequentially
specifically cleave host cell RNA, leading to host cell or
cell death latency [18,19].
Taking this information into account, this work aimed to
propose an anti-COVID-19 therapeutic alternative based on
the CRISPR-Cas13 system, and generate mutations in the virus,
specifically in the sequence of the SARS-Cov2 protein S RBD gene
that It has the function of binding to the cellular ACE2 protein and
that it is the entry site of the virus for its infectivity.
Methods
First, an exhaustive review of the reported sequences of Sars- Cov2 was made, in the databases: NCBI, Google Scholar, Scopus, GeneBank, NIH. With descriptors COVID-19, CRISPR-Cas13, Gen SARS-CoV-2. The gene sequences and spike protein sequences for SARS-CoV-2 were downloaded to FASTA from GeneBank NCBI Reference Sequence: NC_045512.2. To determine the binding points of the virus to the ACE2 receptor, the sequence reported by Walls A, et al. [20] in Cell was taken as a model. With the MEGA software [21]. The sequences were compared, and mutational analysis was carried out. In addition, mutations were made in silico that allowed the generation of stop codons that would prevent the configuration of the virus RNA and make it unfeasible for its duplication and subsequent infection. Multiple couplings of the RBD site and the ACE2 receptor were made to find binding to it and whether or not it was diminished. This was run in PyMol software [22].
Results and Discussion
SARS-CoV-2 is an RNA virus, so the best technology to attack it inside the cell is when it deposits its genetic material in the cell. The appropriate technique presented is CRISPR-Cas13, which attacks RNA and not DNA [23,24]. Based on this, the following strategy was proposed here using its CRISPR technology: This is the sequence of the Virus genome, downloaded in FASTA from GeneBank: NCBI Reference Sequence: NC_045512.2. the genome corresponding to Surface Glycoprotein S. Below, highlighted in yellow is the genome of the virus and the proteins of the virus [25].

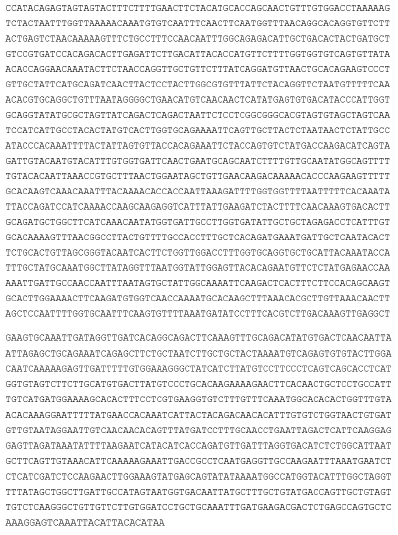
This is the sequence of all the virus proteins, downloaded in FASTA from GeneBank: NCBI reference sequence: NC_045512.2. (In yellow, the genome corresponding to Surface Glycoprotein S).

According to Walls, et al. [20]
The protein sequence has points that are the ones with the highest binding to the ACE2 receptor in the cells of our body. Marked in the figure below with asterisks [20].
When the protein and genomic sequences were compared with the MEGA software, each of the codons corresponding to these binding sites were obtained (marked in red and in bold):

The proposed system is for it to be removed with CRISPR-Cas13, the genome sequence of the virus surface glycoprotein S, within the cell and then replaced by the following sequence with all the binding sites changed by stop codons (the T was changed so that U converts it to RNA, from the original sequence):
Figure 2 shows a coupling between the RBD of the virus with the ACE2 protein. It is appreciated that when the RBD is not mutated there is better coupling with the receptor
Figure 2:
A. RBD spike protein (Right, without mutations) bound to left ACE2.
B. RBD spike protein (Right, with mutations) bound to left ACE2. Created in PyMol.
Conclusion
In this result, some mutations were made in the possible sgRNA
fragments and new mutations could be inserted as stop codons
that would prevent the growth of the virus. This technique can be
worked Ex vivo and, in this way, add to the cells strains sensitive
to the action of our immune system. In this way, the virus will be
unable to infect other cells. There is a truncated coupling when the
protein is mutated.
For more Articles on : https://biomedres01.blogspot.com/

No comments:
Post a Comment
Note: Only a member of this blog may post a comment.