An Early Excessive Release of Cytokines by Ionizing Radiation Exposure in Macrophages Accentuates Inflammatory Disorders
Research Article
Introduction
Cytokine production is an important regulator of immune responses; however, excessive production following radiation exposure accentuates inflammatory diseases and various disorders [1-4]. Inhibition of the excessive release of cytokines or overexpression of their receptors is considered a useful strategy in protecting the effects of radiation [5,6]. Therefore, substantial research has been performed to investigate methods to prevent cytokine-mediated disorders by targeting specific cytokines, including tumor necrosis factor (TNF)-α [1], and transforming growth factor (TGF)-β1 [2,4,7]. However, inhibiting cytokine release or receptor expression might have less of an effect on preventing the induction of disorders in certain tissues characterized by high levels of cytokine production, such as the lung [6]. Macrophages are immune cells that produce cytokines in response to immunerelated insult.
To attain a functional phenotype, macrophages can be recruited and differentiated to an M1 or M2 polarization, with M1 macrophages characterized by the production of proinflammatory cytokines that enhance immune responses [8]. Several reports show that following radiation exposure, carcinogenesis represents a potential long-term consequence, and that the resulting excessive production of cytokines, such as TNF- α, interleukin (IL)-1, and TGF- β1, increases the risk of inflammatory disorders [9,10]. For example, during the treatment of lung cancer, increased levels of TNF- α induce radiation pneumonitis [6], and evidence shows that higher levels of interferon (IFN)-γ during breast cancer treatment might potentiate the induction of inflammatory disorders [11]. Moreover, in the event of radiological accidents resulting in wholebody irradiation, increased levels of cytokine release are related to the presence of autoimmunity associated with thyroid diseases in exposed individuals [12,13]. Therefore, cytokine activation following radiation exposure plays a major role in immune responses resulting from radiation-induced immune insult.
Following radiation exposure, multiple cytokine-related pathways are activated in association with the early regulation of cytokine mRNA transcripts [9]. Among the pathways involved in early regulation of cytokines is the redox-mediated pathway [14], wherein excessive production of cytokines is associated with oxidative stress detected by cytosolic DNA markers, such as zinc finger binding protein-1 (ZBP1) and DNA cross-link repair- 1C (DCLER1C), levels of which are upregulated by activation of IFN-regulatory factors (IRFs) [15]. Previous studies reported that increased levels of these gene transcripts, as well as those of hemeoxygenase-1 and lipocalin-2, are associated with prolonged cytokine production and radiation-induced inflammatory disorders [9,13,14,16,17]; however, a direct relationship between excess cytokine production and the initiation of such disorders has not been established.
In this study, we demonstrated that radiation exposure resulted in cytokine overproduction associated with the differential expression of molecules related to the inflammatory response and cytokine activity according to gene ontology (GO) results. We hypothesized that excessive release of cytokines from macrophages is related to the network of cytokine-related inflammation associated with responses to oxidative stress. Our findings highlighted a relationship between radiation-induced overproduction of cytokines by M1 macrophages and the induction of oxidative stress and upregulated expression of cellular-injury markers.
Materials and Methods
Cell Culture
RAW264.7 murine macrophages were used for this study. The cell line was purchased from the American Type Culture Collection (Rockville, MD, USA) and cultured at 37 °C in 5% CO2 in Dulbecco modified Eagle medium (Gibco, Gaithersburg, MD, USA) supplemented with 10% heat-inactivated fetal bovine serum (Gibco) and 1% Penicillin/streptomycin (Gibco). The cells at passage five to 15 were used for experiments.
M1-Priming of RAW264.7 Cells
Cells were programmed to M1 macrophages, as described previously with slight modifications (20). Briefly, cells were plated in 65-mm dish at a density of 0.5 × 106 cells/plate and primed to M1 macrophages by the addition of 100 ng/mL LPS (Escherichia coli 0127:B8; Sigma-Aldrich, St. Louis, MO, USA) for 24 h, followed by washing twice and treatment with 10 ng/mL IFN-γ (Peprotech Inc., Rocky Hill, NJ, USA) for another 24 h. Macrophages were washed twice and incubated for 12 h with RF-free medium before irradiation.
Flow Cytometry Analysis to Determine M1 Macrophages Lineage
After LPS/IFN-γ treatment, surface-expression markers of M1 macrophages were determined by flow cytometry using a Cytomics FC 500 system (Beckman Coulter, Fullerton, CA, USA). A total of 0.5 × 106 cells was primed and harvested for flow cytometric analysis. Cells were stained with 0.5 mg/mL fluorescein isothiocyanatelabeled anti-mouse F4/80, 0.2 mg/mL of phycoerythrin-labeled hamster anti-mouse CD80, and 0.2 mg/mL Alexa-Fluor rat antimouse DC206 antibodies for 20 min at 4°C in the dark. Cells were analyzed using specialized software (CXP software; Beckman Coulter).
Gamma Irradiation
To avoid the influence of RFs and LPS/IFN-γ during and after radiation exposure, primed RAW264.7 macrophages were further incubated with RF-free media for 12 h and irradiated with a 60-Co gamma cell (MDS Nordion, Ottawa, ON, Canada) at a dose of 2 Gy, 6 Gy, or 10 Gy. After irradiation, cells were incubated at 37°C in 5% CO2 for various times. Harvested cells were either directly used for experiments or stored at the appropriate temperature until later use.
Cell Viability and Cytotoxic Test
Cell viability following IR exposure was measured using a CytoX assay kit (Daeil Lab Service, Seoul, Korea). In each well of a flatbottom 96-well plate, 0.3 × 104 cells/well were seeded for 24 h and polarized to the M1 phenotype, as described. M1-primed cells were exposed to various doses of gamma radiation and incubated for another 12 h. CytoX reagent was then added, followed by incubation for 2 h. Cell viability was assessed using a microplate reader (Tecan, Männedorf, Switzerland) at 450 nm, and cell viability was expressed as the percentage difference from the control at the corresponding dose.
Western Blots
Primed RAW264.7 cells were harvested at an appropriate time point after IR exposure and lysed with radioimmunoprecipitation assay buffer (RIPA; Rockland, Limerick, PA, USA). The lysate was incubated for 20 min in ice and subsequently centrifuged at 13,000 rpm for 15 min at 4°C to remove cell debris, after which protein concentration was determined using a bicinchoninic acid (BCA) assay (Thermo Fisher Scientific, Pittsburgh, PA, USA) according to manufacturer instructions. We performed 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, followed by probing of the membranes overnight at 4°C using primary antibodies against iNOS, COX-2, and glyceraldehyde 3-phosphate dehydrogenase (Cell Signaling Technology, Danvers, MA, USA). The blots were then incubated with a secondary horseradish peroxidase (HRP)- conjugated anti-mouse or anti-rabbit IgG antibody (Cell Signaling Technology) for 1 h at room temperature. Protein bands were detected using an enhanced chemiluminescence (ECL) reagent (Thermo Fisher Scientific).
Cell Preparation for RNA Isolation and PCR Analysis
Total RNA was isolated from IR-treated and untreated primed cells using TRIzol reagent (Thermo Fisher Scientific) according to manufacturer instructions. RNA concentration was assessed using a Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA), and RNA integrity was assessed according to 18S:28S ratio by gel electrophoresis. Samples meeting RNAquality criteria were converted to cDNA using a Labo PassTM cDNA synthesis kit (Cosmo Genetech, Co., Ltd., Seoul, Korea) for further analysis by Real Time PCR and quantitative PCR (qPCR). The qPCR was performed with the SYBR Green PCR core reagent kit (Applied Biosystems; Thermo Fisher Scientific).
Microarray Analysis
Gamma-irradiated and non-irradiated RNA samples were amplified and hybridized using an Affymetrix Mouse Gene 2.0 ST microarray (Affymetrix, Santa Clara, CA, USA) according to manufacturer instructions. After scanning the arrays, differentially expressed probe sets were selected based on 2-fold changes relative to the control. DEGs were analyzed using a heatmap and grouped into early, medium-, and late-response genes based on the time post-IR exposure.
Computational Analysis
The mouse genome informatics database (http://www. informatics.jax.org/) was used to name and group genes into various GO categories based on their biological processes and molecular functions [18]. GO terms exhibiting significant enrichment (p < 0.05) were selected for further analysis. DEGs at the 0.5-h time point and representing biological processes were further analyzed using DAVID (v6.7; https://david.ncifcrf.gov/) to identify associations between GO terms and functional annotations. Pathway analysis was performed using the reactome pathway browser [19]. An FDR between 0.9 and 1 was used as the cut-off for functional-association analysis.
Cytokine Analysis
Cytokine profiles were determined using the mouse cytokine array panel A (R&D System, Inc., Minneapolis, MN, USA). Cells were extracted using RIPA lysis buffer, and protein concentration was determined by BCA assay. Equal amounts of cell lysates were mixed with a cocktail of biotinylated detection antibodies, followed by incubation with the cytokine array membrane according to manufacturer instructions. Streptavidin-HRP and ECL reagents were added to the array membranes to detect bound cytokines. Blots were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA), and intensities were calculated as fold changes relative to the control.
Determination of ROS
We used the 2’,7’-dichlorofluorescin diacetate (DCFDA) cellular ROS kit (Abcam, Cambridge, UK) to detect ROS levels in IR-exposed cells. Briefly, RAW264.7 cells were primed to M1 macrophages and irradiated with various doses of gamma radiation, followed by incubation for another 2 h and labeling with DCFDA according to manufacturer instruction. Tert-butyl hydrogen peroxide was used as the positive control, and absorbance at 485 nm was determined using a microplate reader (Tecan, Männedorf, Switzerland). The mean fluorescence intensity of three replicates was used to calculate changes in DCF fluorescence.
Statistical Analysis
All experiments were performed in triplicate, and data are presented as the mean ± standard error of the mean. Student’s t test was used for comparisons and, when necessary, a p < 0.05 indicated statistical significance.
Results
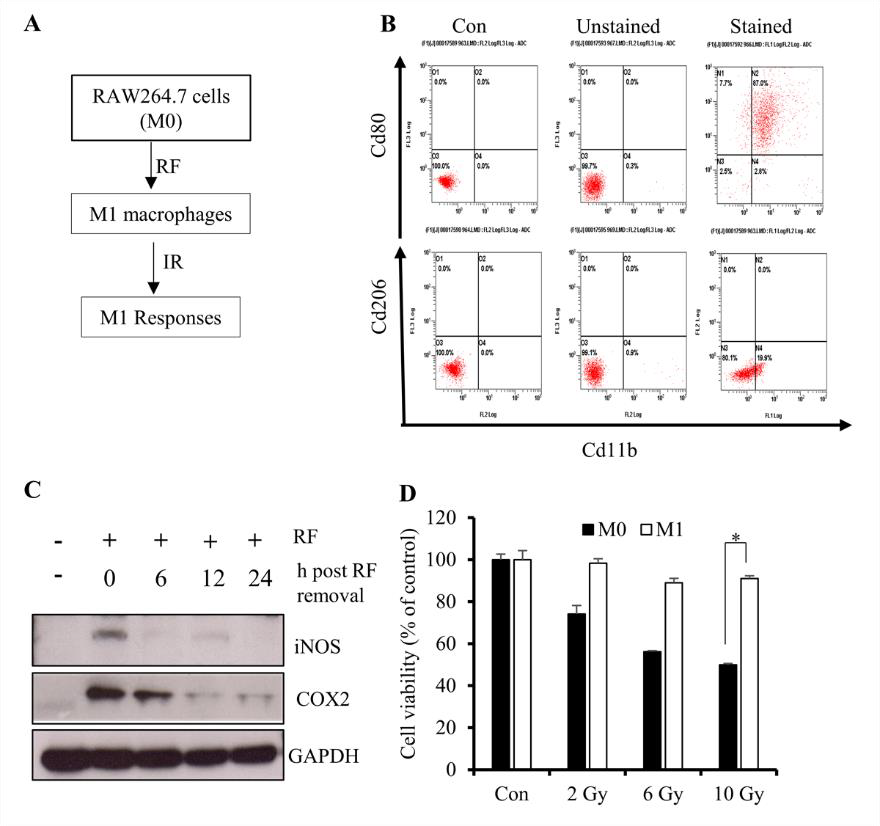
M1-primed RAW264.7 Cells Display Reduced Levels of Proinflammatory Markers Before Exposure to Ionizing Radiation (IR)
Several studies report that monocytes can be primed with several reprogramming factors (RFs) to promote polarization to an M1 [lipopolysaccharide (LPS) and IFN-γ] or M2 (IL-4 and IL- 13) macrophage phenotype [20-22]. Because M1 macrophages are associated with inflammatory cytokine production, we primed RAW264.7 cells in order to investigate signatures associated with cytokine overproduction upon IR exposure (Figure 1A). Flow cytometry results revealed elevated expression of the M1-surface marker CD80 relative to the M2-surface marker CD206 (Figure 1B), suggesting initiation of M1 polarization. M1 macrophages are characterized by elevated expression of inflammatory markers, such as induced nitric oxide synthase (iNOS) and cyclooxygenase (COX)-2, with their levels upregulated at earlier time points following IR exposure [23]; therefore, we considered reduced levels of these molecules as pre-IR markers. Western blot results showed that culturing primed cells in RF-free media returned iNOS and COX-2 to basal levels at 12 h after removal from media containing RFs (Figure 1C).
We then determined whether the differentiated RAW264.7 cells maintain their viability and displayed the appropriate functional responses following IR exposure, finding that >85% of cells remained viable following exposure to 10 Gy of gamma radiation as compared with 100% of non-differentiated cells (Figure 1D). These results suggested that IR exposure at 12-h post-RF removal was a viable experimental protocol for the use of M1-primed RAW264.7 cells.
Figure 1: Differentiation of RAW264.7 cells into M1 macrophages.
A. Schematic description of the priming procedure. RAW264.7 cells were primed to M1 macrophages followed by IR exposure at various doses.
B. Flow cytometric results showing the percentage of the M1 population among primed RAW264.7 cells.
C. Western blot analysis of iNOS and COX-2 levels after 12 h of incubation in the absence of RFs (LPS and IFN-γ).
D. CytoX assay using primed cells and showing the viability of irradiated M1-primed RAW264.7 cells (M1 macrophages) and non-primed cells (M0). *p < 0.05.
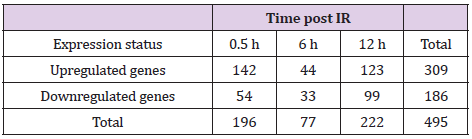
Differentially Expressed Genes in M1-primed RAW264.7 Macrophages Following IR Exposure
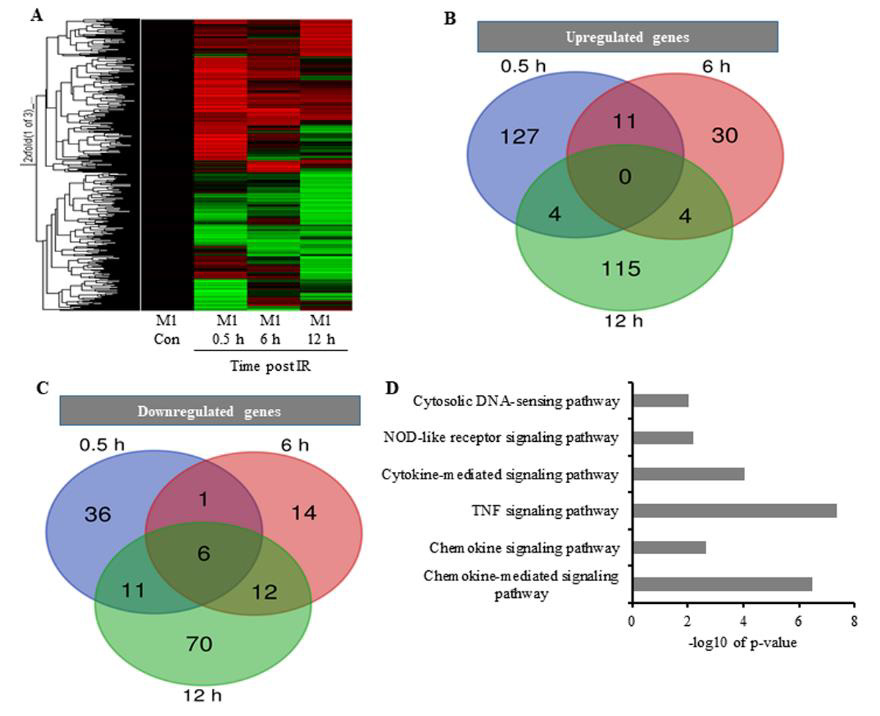
We then investigated changes in gene transcription associated with cytokine overproduction following IR exposure. Cell processes require transcriptional regulation to balance responses to immune insult. Here, primed cells were irradiated and subjected to microarray analysis to identify changes in gene-expression profiles. Differentially expressed genes (DEGs) exhibiting a >2-fold change, a p < 0.05, and a false discovery rate (FDR) <1 were analyzed and grouped according to early, medium, and late responses according to their expression post-IR exposure (Figures 2A-2C; Table 1). Database for Annotation, Visualization, and Integrated Discovery (DAVID) and online analysis tools were used to sort DEGs into functional categories based on their biological process and molecular function (Supplementary Table 1). Microarray results indicate that at 0.5-h post-IR, 142 genes were upregulated and 54 genes were downregulated (Table 1), and that among the 495 genes altered by IR exposure, 11 (2.2%) were exhibited constant upregulation from 0- to 6-h post-IR (Figure 2B) and were associated with the inflammatory response (GO: 0006954), cytokine activity (GO: 0005125), and cellular response to IL-1 (GO: 0071347) (Kappa value > 0.9).
Table 1: Summary of the microarray results.
Note: GO analysis of DEGs identified at various time points post- IR. After priming and IR exposure, ~495 DEGs were clustered and categorized into three groups of early (0.5 h), medium (6 h), and late (12 h) responses.
Figure 2: The effect of IR on the expression pattern of M1 macrophages;
A. Heatmap analysis of microarray results. The 2-fold changes of differentially expressed genes of 10 Gy irradiated macrophages were analyzed by heatmap 0.5 h, 6 h, and 12 h post-treatment. Red indicates upregulated genes, green downregulated genes, and black unchanged genes. Genes were clustered with an agglomerative clustering algorithm into early, middle and late expressed genes.
B. Venn diagram depicting upregulated DEGs and
C. For down-regulated DEGs.
D. DAVID analysis showing functional annotation of biological process pathways for upregulated DEGs.
These GO terms are associated with genes transcribed in response to cellular injury induced by physical agents and increased levels of cytokines released by macrophages. Additionally, the results indicated that earlier upregulated genes were associated with major pathways involved in immune responses post-IR. Reactome and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses showed that most of the DEGs were clustered in pathways associated with TNF, chemokine-mediated, and cytokine-mediated signaling (Figure 2D), all of which are critical to upregulated cytokine production [24]. These results confirmed an association between IR-induced DEGs and potential cytokine overproduction following radiation exposure.
IR-Induced Overproduction of Cytokines in M1-primed RAW264.7 Cells
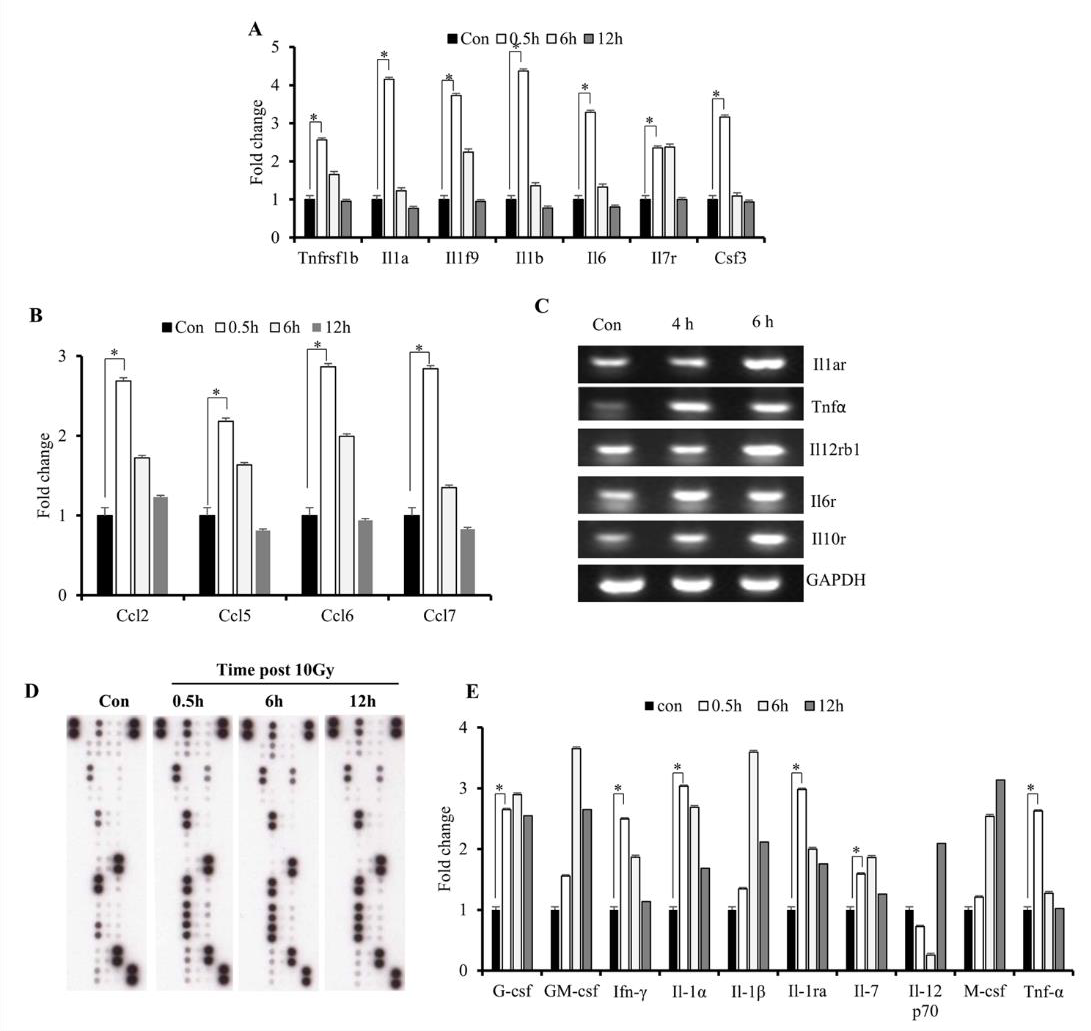
Increased cytokine expression upon IR exposure might suggest involvement of the identified DEGs in the initiation of radiationinduced disorders. Excessive production of cytokines has been associated with various inflammatory disorders [25]; to determine cytokine response to radiation in the immune microenvironment, we investigated levels of early released cytokines both at the mRNA and protein levels. The results showed elevated mRNA levels of Il-1 α / β, Tnf- α, and Ifn-γ (Figures 3A & 3E), as well as C-C motif chemokine ligand (Ccl) 2, Ccl5, Ccl6, and Ccl7 (Figure 3B), at 0.5- h post-IR exposure. Although mRNA levels of these cytokines decreased at 6 h, we found that levels of their receptors remained elevated (Figure 3C). Additionally, we observed sustained elevated expression of TNF-receptor superfamily member 1B (Tnfrs1b), Il- 19, and Il-7 over the course of 6 h (Figure 3A). Furthermore, we found that protein levels suggested similar increases in IL-1 and TNF- α levels at the 0.5-h time point (Figures 3D & 3E), which agreed with a previous study [10]. The results indicated that IR induced excessive production of cytokines immediately following exposure.
Figure 3: Effects of IR on gene expression in M1 macrophages.
A. Microarray analysis of early expression cytokine transcripts at various times following IR exposure.
B. Reverse-transcription PCR analysis of early expression of chemokines and CSFs.
C. Reverse-transcription PCR analysis of cytokine-receptor levels. D, E) Cytokine array analysis of M1-primed RAW264.7 cells exposed to either sham IR or 10 Gy gamma radiation, followed by incubation for the indicated times.
D. Array blots and
E. cytokine protein levels. *p < 0.05.
Changes in the IR-Induced Inflammatory Microenvironment Following Excessive Production of Cytokines
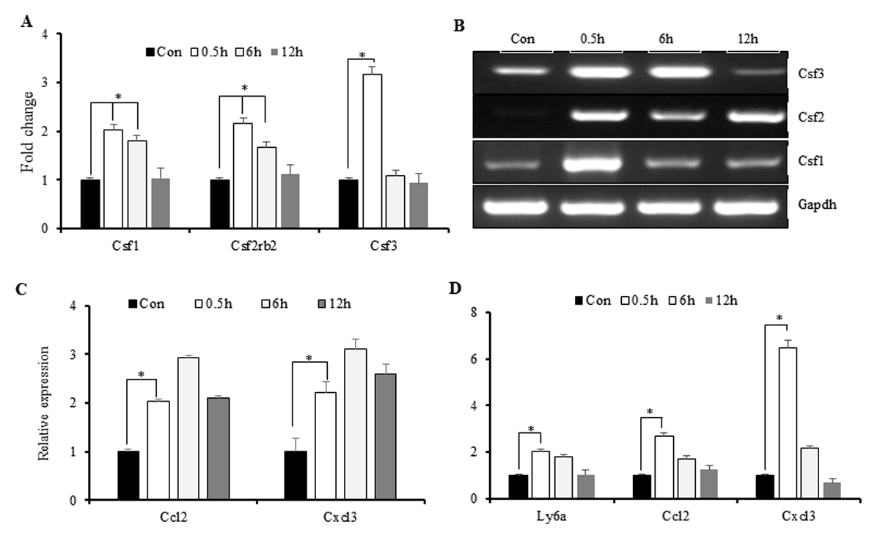
The exposed microenvironment is characterized by elevated cytokine levels, as well as those of colony stimulating factors (CSFs) and chemokines, which support the recruitment of inflamed monocytes to aid the immune response [26]. To correlate this activity with IR-induced increases in cytokine levels, we investigated changes in CSF and chemokine expression following IR exposure. Microarray results showed an increase in Csf-3 levels at 0.5-h post-IR exposure (Figure 4A), with these results confirmed by polymerase chain reaction (PCR). Additionally, we found that elevated Csf-1 and Csf-2 levels were sustained for up to 6-h post- IR exposure, indicating extended recruitment of immune cells to the inflamed microenvironment (Figure 4B). Similarly, we found elevated levels of C-X3-C motif ligand (Cxcl)3 and Ccl2 maintained from 0.5- to 6-h post-IR exposure, indicating increased cytokine expression (Figure 4C). Additionally, we observed increased surface expression of lymphocyte antigen 6 complex, locus A, on M1 macrophages, suggesting their acquisition of an antigenpresentation phenotype associated with cytokine secretion (Figure 4D) and representing a marker indicating monocyte recruitment to the site of injury [27]. These results indicated that M1-primed RAW264.7 cells exhibited macrophage-like behavior involving IRinduced secretion of cytokines.
Figure 4: Immune markers induced by macrophages in response to IR.
A. Microarray analysis of IR-induced expression of CSFs, followed by
B. PCR confirmation.
C. PCR analysis of chemokine expression and
D. markers indicating monocyte recruitment to the inflamed microenvironment. *p < 0.05.
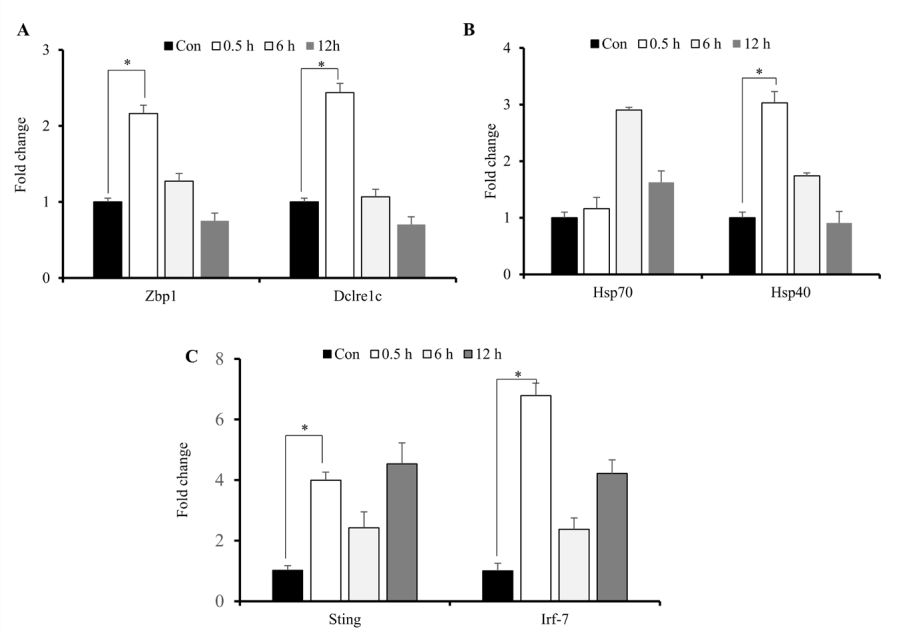
Upregulation of Cytosolic DNA Sensors Following IRInduced Immune Responses
During radiation exposure, initial inflammatory responses are associated with intracellular signaling associated with recognition of and response to damage-associated molecular patterns and free radicals, release of DNA to the cytosol, and increased production of heat-shock proteins (HSPs) and cytokines [15]. Because persistent DNA damage in irradiated cells correlates with prolonged cytokine production which are upregulated by activation of IFN-regulatory factors [25,28], we assessed concurrent increases in cytosolic DNA markers and IR-induced elevations in cytokine expression. We identified significantly increased expression of Zbp1, Dclrec1c, Hsp40, Hsp70, Sting and Irf-7 primarily at 0.5-h post-IR exposure (Figures 5A-5C). These results indicated that IR-induced elevations in cytokine production and release initiated cellular responses stress.
Figure 5: IR-induced increases in cytosolic DNA sensors. Quantitative reverse-transcription PCR analysis of sensors of cytosolic DNA and markers of cellular stress response. A, B, C) Zbp1, Dclrec1c, Hsp40, Hsp70, Sting and Irf-7 mRNA levels after exposure of M1-primed RAW264.7 cells to 10 Gy radiation. *p < 0.05.
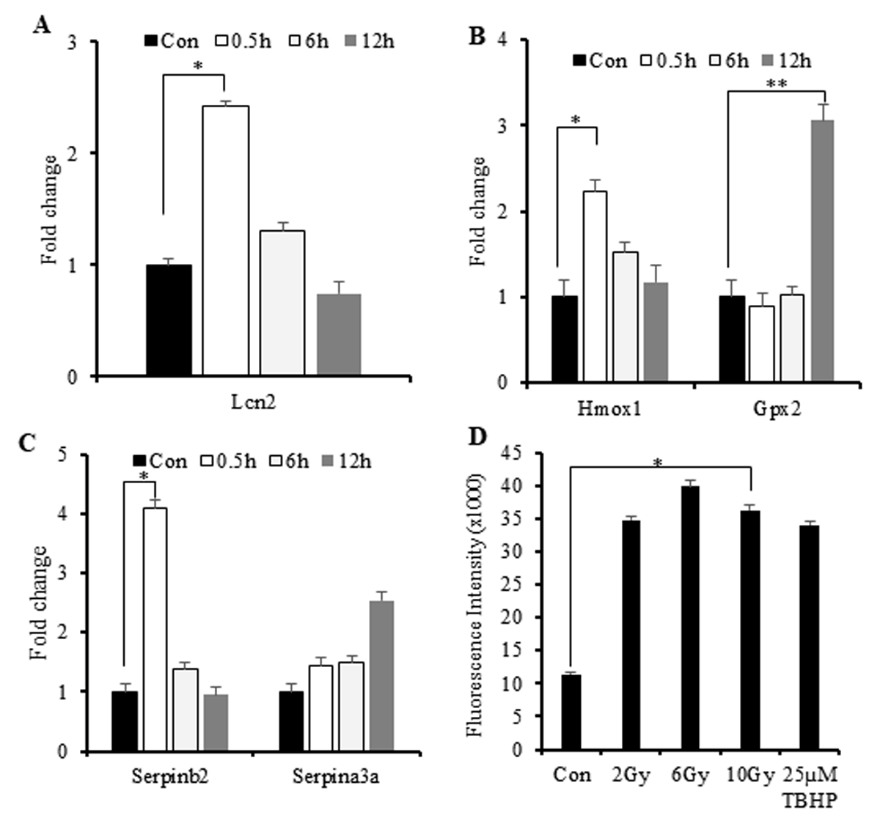
IR-Induced Cytokine Production Results in Elevations in Markers of Oxidative Stress and Cellular Injury
Figure 6: IR-induced increases in the expression of genes involved in the response to oxidative stress and elevated ROS levels.
A. A–C) Quantitative reverse-transcription PCR results showing elevations in Lcn2, Gpx2, Hmox-1, and Serpin mRNA levels.
B. D) ROS levels according to DCFDA assay using irradiated M1-primed RAW264.7 cells. Tert-butyl hydrogen peroxide was used as a positive control. The mean fluorescence intensity of three replicates was used to calculate changes in fluorescence. *p < 0.05; **p < 0.0001.
We then investigated associations between IR-induced cytokine production and increased oxidative stress. IR exposure results in increased levels of reactive oxygen species (ROS/reactive nitrogen species (RNS) primarily generated by mitochondria and associated with cellular injury. To determine whether the IR-induced immune responses can be attributed to cellular ROS/RNS levels, we investigated changes in the expression of genes involved in the response to oxidative stress [9,29]. Lipocalin-2 (Lcn2) is a marker gene associated with radiation-induced cellular injury. Here, we observed increased Lcn2 and heme oxygenase-1 (Hmox-1) expression at 0.5-h post-IR exposure (Figures 6A & 6B), suggesting an early response to radiation injury.
Additionally, we observed late downregulated expression glutathione peroxidase 2 (Gpx2), a scavenger of free radicals in response to oxidative stress (Figure 6B). This finding and the observed upregulation of Hmox-1 suggested macrophage-specific increases in anti-inflammatory activity in response to oxidative stress. To address this phenomenon, we investigated the expression of serine proteinase inhibitor (serpin) family genes (Serpinb2 and Serpina3a) involved in the cellular response to environmental stress associated with inflammation [30]. The results indicated early upregulation of Serpinb2 at 0.5-h post-IR exposure and late upregulation of Serpina3a at 12-h post-IR exposure (Figure 6C), suggesting responses to inflammatory stress induced by radiation. Subsequent assessment of ROS in irradiated macrophages revealed elevated ROS at 2-h post-IR exposure (Figure 6D). These findings suggested macrophage exposure to IR rapidly induced (within 0.5 h) increased expression of genes related to the response to oxidative stress, which promoted activation of cytokine production.
Discussion
We performed computational and microarray analyses to investigate signatures associated with IR-induced excessive production of cytokines in M1 macrophages. Microarray results identified 495 IR-induced DEGs, of which 11 (2.2%) were constantly upregulated for up to 6 h (Figure 2B) and associated with GO terms involving the inflammatory response and cytokine activity. Previous studies reported that early radiation-induced expression of inflammation-related genes were related to CpG islands in the promoters of cytokine genes [9,31]. Additionally, we found that few of the early upregulated cytokines maintained their expression at 6-h post-IR exposure (Figure 3), suggesting that cytokine overproduction occurred early in the response to radiation. To elucidate cytokine overproduction by IR, several molecules were investigated. We associated the detection of cytosolic DNA markers with increased levels of cytokine production.
The expression of Zbp1 and Dclrec1c the cytosolic DNA markers were reported to be associated with the inflammatory responses and cellular injuries [15]. Furthermore, several reports associated the emergence of ZBP1 as a central regulator of inflammatory responses [32]. DCLREC1C is involved in repairing IR-induced damage to double-stranded DNA and might be required for processing prior to re-ligation [28] whereas ZBP1 is an important detector of cytosolic DNA. We strong consider the induction of these genes with increased cytokine levels. Here, we found that Zbp1 and Dclrec1c expression was upregulated in response to IR exposure, suggesting a correlation between the inflammatory response and radiation-induced DNA damage. Furthermore, we ascertain the increased levels of cytokines with induction of key cytokines. IL1 and TNF- α are major cytokines frequently induced early in the response to radiation exposure (as indicated in Figure 3E), and their induction drives subsequent cytokine expression and release [6]. For example, during lung cancer treatment, increased levels of TNF- α induce radiation pneumonitis [6], with a similar outcome reported in association with elevated IFN-γ levels following radiation-mediated breast cancer treatment [11].
Additionally, the release of these cytokines is accompanied by the upregulated production of chemokines and other growth factors in the inflamed microenvironment, with subsequent interactions between inflammatory factors necessary for the immune response; however, excessive responses are related to the onset of autoimmune diseases, which remain a therapeutic challenge. Previous studies reported increased levels of radiationinduced ROS and RNS during the inflammatory response [33,34] and involved in elevated expression of cellular-injury markers, such as Lcn2. Our results supported associations between IRinduced inflammatory responses and elevations in ROS/RNS levels [35]. We found increased ROS levels in IR-exposed macrophages at 2 h, indicating activation of the acute phase of the inflammatory response according to increased ROS-scavenging activity [36]. Furthermore, elevated Lcn2 expression suggested that oxidative stress represented a key step in the cellular response to radiation, with this supported by upregulated levels of Gpx2, Hmox-1, and Serpin mRNA.
In conclusion, we showed that exposure of M1 macrophages to radiation resulted in cytokine overproduction. Our results indicated that secondary exposure to cellular stress induced by radiation further elevated cytokine production within 0.5h of exposure, suggesting that the upregulated immune response was augmented by secondary signaling associated with cellular stress. These results provided insight into relationships between radiation-induced cytokine production, and oxidative stress, and cellular injury.
| For more Articles on : https://biomedres01.blogspot.com/ |



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.