Cloning, Eukaryotic Expression Vector Construction and Tissue Expression Analysis of Mir-133b in Pigs
Introduction
Non-coding RNAs refer to RNAs that cannot encode proteins, among which microRNAs (miRNAs) are a class of small singlestranded RNAs with a length between 20nT-25nt. As an endogenous and highly conserved ncRNA, miRNAs play a role in muscle development and fat metabolism of animals [1], disease regulation [2] mainly playing a role in post-transcriptional regulation. In recent years, there have been more studies on miRNAs in domestic animals, especially domestic pigs, which are economic animals. Studies have reported that there are 10 significantly different miRNAs in muscle tissue between Laiwu pigs and white pigs [3]. MiR-204 regulates piglets’ resistance to Clostridium perfringens type C infection [4]; In the study of Meishan pigs, it was found that miR-208b can affect the transformation of different muscle fiber types by targeting the inhibition of Mettl8 [5].
Initially, miR-133b was thought to be specifically expressed in muscle with a tissue-specific expression pattern, and highly expressed in the heart and skeletal muscle, and playing a key regulatory role in muscle development and remodeling [6]. It has been reported that miR-133b can be used as an indicator for early diagnosis of diabetic nephropathy [7] and can regulate the proliferation process of gastric cancer cells [8]. MiR-133b-siRT1- β-catenin axis can be involved in sporadic gliomas [9]. The function and target genes of miR-133b have been extensively studied, and the study on pigs is worth exploring. In this paper, the tissue expression profile of miR-133b was drawn and the eukaryotic expression vector pEGFP-N1-miR-133b was constructed, laying a theoretical foundation for further exploring the mechanism of miR- 133b in pigs.
Materials and Methods
The Experimental Materials
Blood and tissue samples, pEGFP-N1 eukaryotic expression vector and 3T3-L1 cells of Du adult pigs were stored and provided by College of Animal Science and Technology, Guangxi University. Blood DNA extraction kit, plasmid extraction kit, endotoxic plasmid extraction kit, gel recovery kit, miRNA reverse transcription kit, restriction enzymes BamH Ⅰ and Hind parabola, pMD19-T vector, T4 ligase, Trizol, etc. were purchased from Baori Medical Technology (Beijing) Co., Ltd.DH5α competent cells were purchased from Beijing Quansi Gold Biotechnology Co., Ltd. Lip3000 transfection reagent and fetal bovine serum were purchased from Thermo Fisher Technologies. PBS, DMEM and Trypsin were purchased from Beijing Solibol Technology Co., Ltd.
Primer Design and Synthesis
The precursor sequence and flanking sequence of miR-133b were searched in the NCBI and UCSC websites, and the cloning primers of miRNA (miR-133BF1, R1) were designed using Oligo 7.0, in which the underlined sequence was the enzyme digestion sequence. The upstream of miR-133b specific quantitative primer was the sequence itself, and the downstream was the general primer. U6 was selected as the internal reference gene, and the primer sequences were shown in Table 1. All primers were synthesized by Nanjing Nuvezan Biotechnology Co., Ltd.
Total RNA Extraction and cDNA Synthesis
Total RNA was extracted from porcine heart, liver, spleen, lung, kidney, dorsal muscle, subcutaneous fat, abdominal fat and other tissues by Trizol method. RNA with OD260nm/280nm value ranging from 1.8-2.0 was reverse transcribed by miRNA and cDNA was synthesized. The 3T3-L1 cells were washed with PBS for three times, then Trizol lysate was added to collect the cells, and the RNA extraction and reverse transcription methods were the same as the tissues.
Cloning of miR-133b
Blood DNA extraction kit was used to extract DNA from the whole blood of Dugang pigs, and it was used as a template for amplification of miR-133b precursors and part of the flanking sequences. The PCR reaction system was 10 μL, of which Ex Taq PCR Mix was 5 μL, both upstream and downstream primers were added 0.5 μL, DNA template was 1 μL, and RNase free H2O was added to the complement to 10 μL. The PCR amplification conditions were denaturation at 98℃ for 10s, annealing at 55℃ for 30s, and extension at 72℃ for 30s, with a total of 35 cycles. The PCR products were detected by 1.5% agrose-gel electrophoresis. After the target strips were recovered and purified, they were attached to pMD19-T vector overnight at 4℃, transformed into DH5α capable cells and coated on LA plate containing ampicillin. After 12h, a single positive clone colony was selected for expansion culture, and the bacterial fluid was identified by PCR. The bacterial fluid with the correct band size was sequenced by Shanghai Sangong.
Construction and Identification of the Eukaryotic Expression Vector of n1-miR-133b
A small amount of plasmid was extracted from the bacterial fluid with correct sequencing results. The pEGFP-N1 vector and pMD19- t-miR-133b recombinant plasmid were digested with BamH Ⅰ and Hind anion, and the bands were detected by 1.5% AGAR gel electrophoresis and the gel was recovered. After that, the purified target fragment and N1 were ligated overnight with T4 ligase at 4℃ without load. After the ligation products were transformed, coated, selected and shaken, PCR identification and sequencing were performed on the bacterial fluid. The product with correct sequencing results was expanded to 6ml, and then the endotoxinfree plasmid was extracted to obtain the eukaryotic expression vector of n1-miR-133b, which was stored in a refrigerator at -40℃.
Verification of Overexpression of miR-133b in 3T3-L1 Cells
The resuscitated 3T3 cells were cultured in 35mm culture dishes and inoculated in 6-well cell culture plates when the cells grew to 90%. When the degree of cell confluence reached about 70%, the empty plasmid pEGFP-N1 and the recombinant plasmid n1-miR-133b were transfected into 3T3 cells using LIP3000 transfection reagent, with 3 replicates for each. The expression of green fluorescence was observed under an inverted microscope 48 hours after transfection and photographed.
Real-Time Quantitative PCR (qPCR)
The cDNA of different tissues and cells was used as the template, and U6 was used as the internal reference gene for qPCR verification. The reaction system was as follows: TB Green Ⅱ 5.0 μl, upper, lower primers 0.3 μl each, cDNA template 2.5 μl, RNASEfree H2O 1.9 μl, and each system was repeated with 3 holes. PCR amplification procedure: pre-denaturation at 95℃ for 30s; 45 cycles of amplification were performed at 95℃ for 5s and 60℃ for 30s to read fluorescence signals. At 65℃, increase by 0.5℃ to 95℃ every 5s, and read the melting curve. The relative expression level of miR-133b was calculated by 2-ΔΔ CT method. Data were analyzed by SPSS 9.4 for one-way ANOVA, P<0.05 indicates significant difference, P<0.01 indicates a significant difference.
Results and Analysis
Expression Analysis of miR-133b Tissue
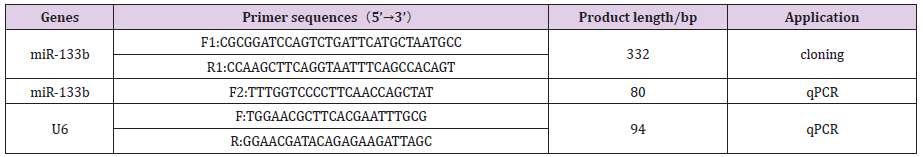
The expression levels of miR-133b in heart, liver, spleen, lung, kidney, dorsal muscle, subcutaneous fat and abdominal fat of Duroc pigs were detected by real-time quantitative PCR, and the results were shown in Figure 1. The expression abundance of miR- 133b was the highest in the muscle tissue, followed by the heart, which was significantly higher than other tissues (P<0.01). It can be inferred that miR-133b plays an important regulatory role in muscle, which is consistent with the characteristic of tissue-specific expression of miRNA (Figure 2).
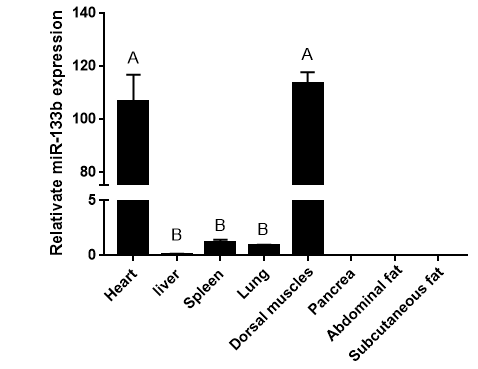
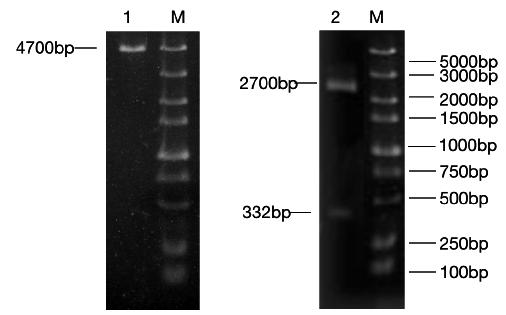
Figure 2: RT-RCR products of miR-133b and its flanking sequence.
Note: M: Marker1; 1: RCR products of miR-133b and its flanking sequence.
RT-PCR Amplification of miR-133b
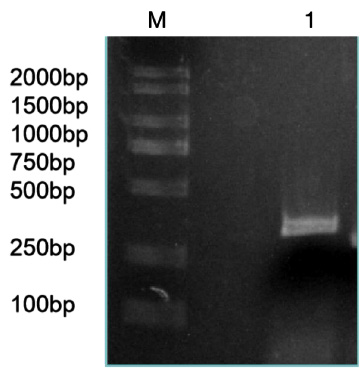
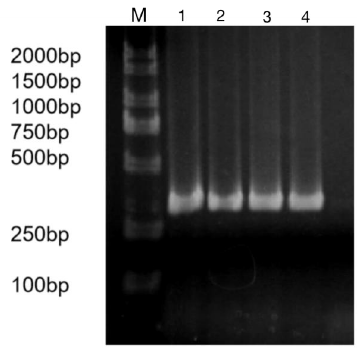
The precursor and part of the flanking sequence of miR-133b were successfully amplified using the DC-grown pig gDNAs as the template. The electrophoresis target band was single and bright, with a size of about 300bp consistent with the expected fragment (Figure 2). RT-PCR identification of bacterial fluid also showed clear bands of the same size (Figure 3). Sequencing results of Sangon Bioengineering (Shanghai) Co., Ltd showed that miR-133b was successfully amplified with a fragment size of 332bp.
Figure 3: Bacteria liquid PCR products of miR-133b.
Note: M: Marker1; 1~4: Bacteria liquid PCR of miR-133b.
Verification of Recombinant Plasmid pEGFP-N1-miR- 133b
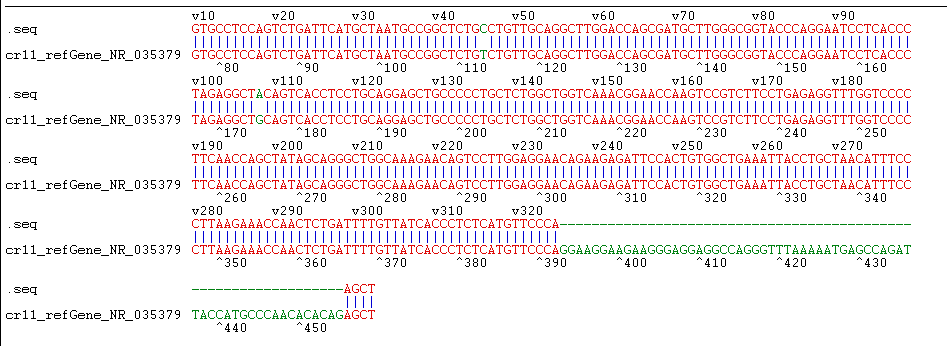
The constructed eukaryotic expression vector of n1-miR- 133b was identified by double enzyme digestion, and two clear target bands with correct size and position were obtained by RTPCR amplification (Figure 4). The fragment size of miR-133b was 268bp, indicating that the vector of n1-miR-133b was successfully constructed. The sequencing results of the recombinant plasmid vector were compared with the sequences provided by the NCBI website, and the results showed that the homology between miR- 133b of Ductus giant pig and the published seed sequences reached 100% (Figure 5). 1 is N1 plasmid double digestion, 2 is the result of pMD19T-MI-133B double digestion.
Figure 4: N1,pEGFP-N1-miR-133b recombinant plasmid double digestion and validation.
Note: M: Marker1;1: Double digestion of N1 plasmid 2:PCR products of pEGFP-N1-miR-133b.
The Recombinant Plasmid pegfp-n1-mir-133b was Transfected into 3T3 Cells
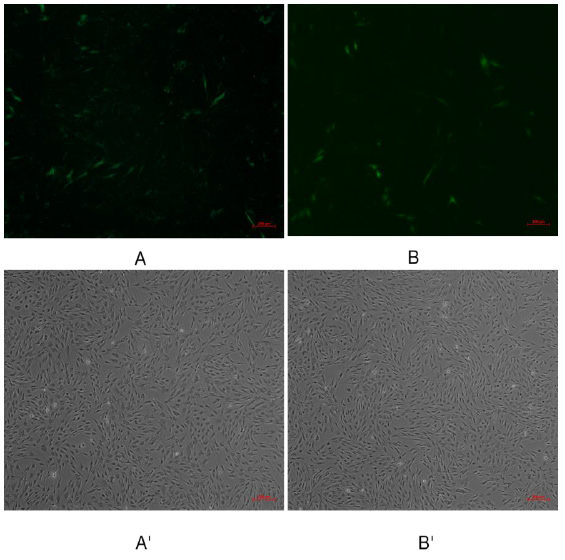
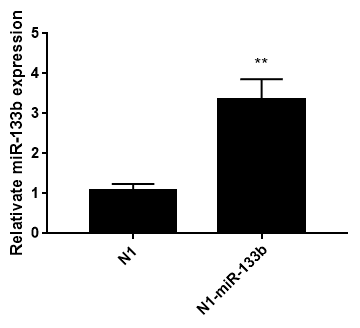
Pegfp-n1-mir-133b eukaryotic expression vector and pEGFP-N1 empty vector were transfected into 3T3-L1 cells respectively. After 48 hours, green fluorescence expression was observed in 3T3-L1 cells under fluorescence inverted microscope (Figure 6), which proved that the recombinant plasmid could be expressed in 3T3 cells. The expression of miR-133b in 3T3 cells was detected by qPCR. The results showed that the relative expression level of pegfp-n1-mir-133b eukaryotic expression vector transfection group was significantly higher than that of empty vector group (P < 0.01) (Figure 7).
Figure 6: Green fluorescence results of 3T3 cells transfected with recombinant plasmid pEGFP-N1-miR-133b.
Note: A, B were fluorescence of transfection withpEGFP-N1-miR-133b, pEGFP-N1 respectively; A’, B’were transfection with
EGFP-N1-miR-133b, pEGFP-N1 under under the common light respectively.
Figure 7: Relative miR-133b expression in 3T3 cells after transfecting 48h.
Note: Date represent means ± SD; ** represented extremely significant difference (p<0.01)
Discussion
MiRNA can regulate a large number of target genes independently by not completely complementary with specific mRNA, and participate in different cell signaling and metabolic pathways, as well as physiological and biochemical pathways such as cell proliferation, differentiation and apoptosis [10]. The development of animal muscle tissue is affected by a variety of regulatory factors, including myogenic regulatory factors MyoD, Myf5, MyoG;Pairing box factors Pax3, Pax7, etc. In recent years, more and more people pay attention to the research of noncoding RNA in muscle development, including Long non-coding RNA(lncRNA), circular RNA (circRNA) and miRNA [11]. MiRNA mainly inhibits its translation by binding the seed sequence (2-8 nucleotides from the 5 ‘end) to the target mRNA. MiR-1, miR-133, mir-486 and miR-206 are collectively referred to as muscle specific miRNAs, which are highly conserved in muscle tissues of different species [12]. Mir-486 participates in IGF-1 / Akt / mTOR signaling pathway to regulate skeletal muscle cell size, and also acts as a signal molecule of myogenic inhibitor [13]. After knocking out miR- 206 in C2C12 cells by CRISP-CAS9 technology, it was found that muscle development was slow, and ADAM19, BGN, Cbx5, Smarce1, and Spg20 were identified as its direct targets [14]. MiR-1 can promote muscle development by inhibiting the post transcriptional expression of HDAC4 in goats [15].
This study found that miR-133b plays an important role in muscle development. In disease treatment, miR-133b can reduce the harmful effects of Duchenne muscular dystrophy [16]. It can alleviate atherosclerosis by regulating vascular smooth muscle [17]. In livestock production, miR-133b can regulate the proliferation of bovine skeletal muscle satellite cells by targeting SP1 gene [18], and together with miR-1a and miR-206a to regulate muscle development of Tongcheng pig and landrace pig [19]. In this study, we constructed the tissue expression profile of Duroc Landrace pigs, and found that miR-133b was the highest expressed in dorsal muscle and myocardium, indicating that miR-133b may be related to muscle development. By constructing miR-133b overexpression vector, qPCR detection showed that the expression level of pegfpn1- mir-133b was significantly higher than that of the control group, indicating that miR-133b has a certain role in animal muscle development. These results indicate that miR-133b is expressed in 3T3 cells and specifically expressed in different tissues of Duroc Landrace pigs. This study provides a theoretical basis for further exploring the mechanism of miRNA in pigs and opens up a new way to improve the quantity and variety of pork.
For more Articles on : https://biomedres01.blogspot.com/



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.