Ligand-Based Virtual Screening for the Inhibitors of Monoamine Oxidase B
Introduction
Monoamine Oxidases
Monoamine oxidase enzymes are central to the normal functioning of brain by playing a key role in neurotransmitter metabolism thus involved in some neurodegenerative diseases including Parkinson’s disease. Existence of isoforms monoamine oxidase A and B was established after separate gene sequences encode two enzymes. These isoforms are differentially expressed with MAO-A expressed in human placental mitochondria and MAO-B in human platelet mitochondria [1]. MAO plays an absolute role in the deamination of neurotransmitters thus controlling their level and concentration in the nervous system consequently linked to the basic pathological signs of neurological disorders. With the know-how of crystal structure of MAO enzyme, it becomes a major drug target and enables ligand designing [2]. MAO are bound to the surface membrane of mitochondria, flavoproteins causing the oxidative deamination of neurotransmitters. Role of these enzymes [3] in the catabolic processes of essential brain amines like dopamine and phenylethylamine causing an intensification of the neurodegeneration due to the process of oxidative stress is the important common sign of Parkinson’s disease [4].
Tissue Occurrence
Two isoforms vary in the level of presence from tissue to tissue mostly with regional differences in the brain. Striatum and hypothalamus are regions with highest level of activity along with cerebellum and neocortex showing low levels of activity. Main function of this enzyme is to provide protection to the body, oxidation of amines from the blood avoiding their entrance into the circulation. MAO-B acts as a metabolic barrier in the blood brain barrier and shields neurons from exogenous amines, ends the action of amine neurotransmitters regulating their intracellular level [5]. MAO-B is present in significant levels in the astroglial cells acting as a biomarker for the crucial process of astrogliosis, which is involved in the mechanism of the Parkinson’s disease providing a positive correlation between MAO-B and astrocyte proteins. In case of MAO-A an increased level was seen in substantia nigra in disease condition [6].
Mechanism of Action: Initially a number of mechanisms of action were proposed. MAOB forms the agent responsible for Parkinson’s disease 1- methyl-4-phenyl-pyridinium which is a neurotoxin from the 1-methyl-4- phenyl-1, 2, 3, 6-tetrahydropyridine (MPTP). Both enzymes have covalently bound cofactor FAD cofactor which is attached to the cysteine. A hydrophobic substrate binding domain along with a membrane binding domain. These enzymes exist as dimers in the solution form. Notable difference in structure of isoform A and B substrate binding sites exists primarily in the amino acid residues.
Inhibitors of monoamine oxidase enzyme increase the monoamine neurotransmitter availability, decreasing the production, low levels of neurotoxins formation, initiation of pro survival genes leading to a neuroprotective effect (Figure 1) [7].
Structure
Monoamine oxidase inhibitors not just act by enhancing the catalytic process but are created by targeting specific residues as imidazoline present in the entrance cavity that is also an important component in determining ligand specificity [9]. Inhibition is achieved by interaction to the coil like structure near to the membrane surface and two cavities, entrance and active providing a gateway [3].
Drug Generations
Thus, these inhibitors help in easing some of the symptoms including motor complications. This widely established role motivated researchers all around the world to design new research techniques [10]. Seligiline, initially an anti-depression drug is the first-generation drug in the treatment of Parkinson’s. Rasagiline came up next with increase in specificity and capability. These drugs can be used in different ways according to the progression of the disease symptoms. In such case a monotherapy or combinational treatment can be recommended. Another greatly investigated inhibitor is safinamide known as an ideal, well tolerated and safe inhibitor [11]. All of these inhibitors basically influence positively to the level of neurotransmitters in the brain protecting them from depletion [12].
Deamination
Among both the monoamine oxidase enzymes isoform B is responsible for more than 80% of the total enzyme activity in brain especially in regions like glial cells, histaminergic and serotonergic neurons. Other than brain it breaks down dopamine and other amines in liver. Scholars revealed that monoamine oxidase B activity correlates to the aging appearing to show more activity in neurodegeneration and highly expressed in astrocytes. Thus, as the age goes on, reduction in dopamine and increase in is expected. Rise in the concentration level of stimulates signaling cascades for apoptosis in cell. Evading the cells from this bio activation can defend from oxidative damage [13].
Investigations led to deep insight into the MAOB structure that resulted in showing hydrodynamic properties. These studies indicate the possibility of perceiving MAOB as an accurate biochemical imaging marker for neurodegeneration [14]. Also, in regard to Parkinson’s disease MAOB is specially involved in pathogenesis of disease by producing reactive oxygen species and neurotoxins such as 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine [6]. Other than this human MAOB enzyme is seen to differ from rat MAOB in terms of specificity towards dopamine and some other substrates. This difference is regarded as an attribute of kinetic behavior of enzyme [15].
Virtual Screening
An effective computer-based strategy, virtual screening (VS), has been used for identification such promising compounds which binds with known structure’s molecular target.
Material and Methods
In-Silico Study
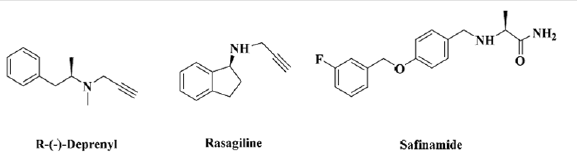
Library Retrieval: Based on published research work we selected three compounds namely selegiline, rasagiline and safinamide for library retrieval of related compounds that can also act as inhibitors. For this purpose, ZINC database was employed that contains over 21 million commercially available, purchasable and ready to screen compounds. Structural similarity search feature of ZINC for rasagiline and safinamide assembled a total of 456 compounds. After downloading from the servers, the compounds w was drawn in MOE builder. All 456 compounds retrieved from zinc database were drawn using this software and saved in cdx file format.
Pharmacophore Generation: Pharmacophore generation is an important technique for recognizing the potential ligands. Basically, a pharmacophore is an orientation in 3D of the functional group of the molecule that plays role in the interaction during binding with the protein. MOE (Molecular Operating Environment) pharmacophore tool was used to generate model for further virtual screening of our data set that comprised of 456 compounds from ZINC database along with dataset of six classes of synthetic compounds. Pharmacophore model created was used to evaluate the database and further filter our dataset to hit compounds. Pharmacophore Query Editing and Searching by MOE Tools: The EHT pharmacophore scheme is based on a semi-empirical approach using Extended Hueckel Theory (EHT) for generating pharmacophore annotations and features. The EHT approach takes into account ligand resonance and electron withdrawing effects and consequently the pharmacophore features generated through the EHT scheme are sensitive to non-standard interactions, such as C-H and halogen bond interactions, during pharmacophore screening.
Molecular Docking: To further refine the shortlisted compounds from pharmacophore analysis, interacting behaviour of these compounds was inspected through molecular docking studies. LeadIT from BioSolveIT, GmbH Germany was used for docking. For this purpose, protein data bank was used to download the crystallographic structure of human monoamine oxidase B (ID: 2V5Z at 1.7 Å). Since MAOB exists as a homodimer so before initiating the docking techniques chain B was removed completely leaving behind just chain A. coenzyme flavin adenine dinucleotide with the MAOB was kept in oxidised form. Prepare receptor utility or load of Lead It software was used for uploading the receptor. Binding site conformation was set at 8.5 Å spacing and docking was initiated using FlexX utility along with default docking parameters. Cognate ligand safinamide was re-docked inside the active site and RMSD value was noted. Ligand-protein complex with lowest free biding energy values were selected for further analysis [16-21].
Visualization: As a result of docking the 3D putative binding modes were visualized using Discovery Studio visualizer v4.
Result and Discussion
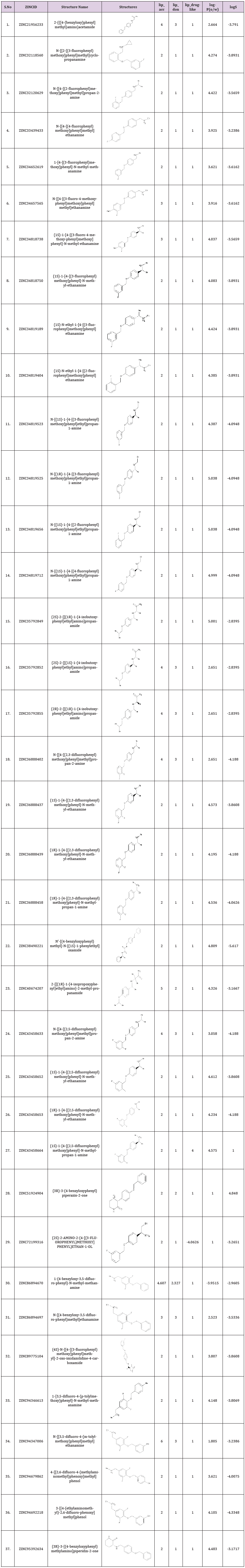
The current studies was based on the identification of potent scaffold from several known inhibitors of monoamine oxidase and marketed inhibitors of MAO-B namely selegiline, rasagiline and safinamide were retrieved from Zinc database and database was prepared. Based upon the pharmacophore, screening of potential inhibitors of MAO-B downloaded from databases were done. Different similarity searching on the basis of structures of rasagiline and safinamide assembled a total of 456 compounds from Zinc database. Out of these 456, 37 compounds were chosen, and filter was applied for the generation of pharmacophore method. The resultant binding interactions and binding energies inside the active pocket were monitored and reported. The lowest binding energy and highest binding interactions were established for ZINC ID 89775104 as -12.43. ADME properties of all the 37 compounds were predicted and physicochemical parameters after the generation of pharmacophore was build (Table 1).
Table 1: ADME properties of all the 37 compounds were predicted from the pharmacophore based screening.
Conclusion
The inhibition of monoamine oxidase enzyme suggests the significant target for the regulation of depression and Parkinson’s disease. The current studies were based on the identification of potent scaffold from several known inhibitors of monoamine oxidase and marketed inhibitors of MAO-B namely selegiline, rasagiline and safinamide were retrieved from Zinc database and database was prepared. Based upon the pharmacophore, screening of potential inhibitors of MAO-B downloaded from databases were done. Different similarity searching on the basis of structures of rasagiline and safinamide assembled a total of 456 compounds from Zinc database. Out of these 456, 37 compounds were chosen, and filter was applied for the generation of pharmacophore method. The resultant binding interactions and binding energies inside the active pocket were monitored and reported. The lowest binding energy and highest binding interactions were established for ZINC ID 89775104 as -12.43. ADME properties of all the 37 compounds were predicted and physicochemical parameters after the generation of pharmacophore was build.
For more Articles on: https://biomedres01.blogspot.com/



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.