Biostatistical Analysis on Anti-breast Cancer Drug Screening
Introduction
Breast cancer is one of the most common malignant tumors in women, and a malignant tumor occurring in ductal epithelium of the breast. Estrogen is involved in the growth and differentiation of mammary epithelial cells in hormone dependent tumors. It plays an important role in the occurrence and development of breast cancer [1]. Estrogen mainly acts through the estrogen receptor expressed in the nucleus, that is, by binding with estrogen receptor (ER) to form a complex [2]. Research shows that ERα is expressed in normal breast epithelial cells less than 10% but expressed in breast cancer cells around 50%-80%. ERα has become an important target of endocrine therapy for breast cancer [3]. Currently, antihormone therapy is commonly used in breast cancer patients with ERα expression, which controls estrogen levels through regulating estrogen receptor activity. ERα mediates the E2 up regulation of PI3K/Akt signaling pathway and promotes cell proliferation [4]. Compounds that can antagonize ERα activity may be candidates for treatment of breast cancer. For example, tamoxifen and renoxifene are the ERα antagonists for clinical treatment of breast cancer [5]. In order to screen potential active compounds, a potential compound model is usually established to collect compounds and bioactive data by targeting the specific estrogen receptor subtype targets associated with breast cancer. The quantitative structureactivity relationship (QSAR) model of compounds was constructed with the biological activity descriptor as the independent variable and the biological activity of compounds as the dependent variable. The model was used to predict the new compound molecules with good biological activity or guide the structural optimization of existing active compounds. A compound that wants to become a candidate drug, besides having good biological activity (here refers to anti breast cancer activity), also needs to have good pharmacokinetics and safety in human body. It is called ADMET property, including absorption, distribution, metabolism, excretion and toxicity. When determining the biological activity of a compound, it is also necessary to consider its ADMET properties as a comprehensive consideration. In this paper, the coupling degree between bioactivity descriptor and ER activity is verified by BP neural network. After determining that the screened bioactivity descriptors can indeed affect ERα activity to a great extent, the ADMET property of bioactivity descriptors is further verified.
Overview of BP Neural Network
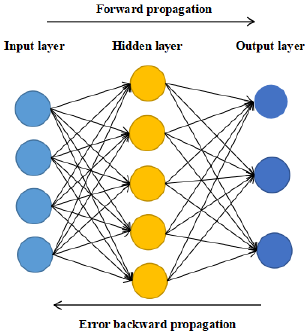
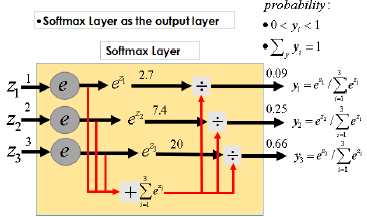
Artificial neural network is widely used in pattern recognition, function approximation and so on. BP neural network is a multilayer feedforward network simulating human brain. It has good adaptability and training ability, belongs to nonlinear dynamic system, and including two processes: forward propagation of information and back propagation of error. BP neural network consists of three parts: input layer, hidden layer and output layer. The input layer receives the input information, and then transmits the information to the hidden layer. The hidden layer analyzes and processes the data. Finally outputs acceptable information through the output layer. This information is continuously corrected through the reverse propagation of error, which can make full use of the coupling between data. BP neural network shows excellent accuracy in many fields. Therefore, this paper selects neural network as the main prediction method. Whether it is regression network or prediction network, the setting of the hidden layer and the number of hidden nodes of the network is very important. Too few hidden layers and hidden nodes will lead to less data information that the neural network can process, resulting in low prediction accuracy, and too many hidden layers will lead to overfitting of the model. There is no general calculation formula for the setting of the optimal number of hidden nodes. It is more based on the empirical formula or changing the number of hidden nodes to continuously train the model to find the number of hidden nodes with the smallest error [6-8]. Basic structure diagram of BP neural network is shown in Figure 1. The activation function of BP neural network usually uses softmax function to give corresponding weight to each node and transfer information between nodes in the network. In addition, there is an offset weight in the propagation of each layer of network, which is an additional constant of SoftMax function. In the model training, the gradient optimization algorithm (Adam algorithm) is used to optimize the model to obtain the best results [9].
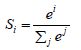
Its operating principle is shown in Figure 2.
Adam Algorithm:
Initialize 1st, 2nd moment vector and timestep:
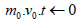
do while:
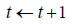
Computing the gradient:
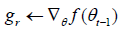
Update biased first moment estimate:
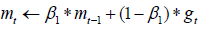
Update biased second moment estimate:
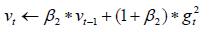
Compute bias-corrected first moment estimate:
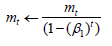
Compute bias-corrected second moment estimate:
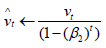
Update parameters:
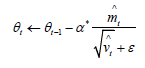
Where α is the step length, β ;β ε [0,1] is the momen estimation of exponential decay rate, and f(θ) is the random objective function of parameter θ. Adam algorithm will be used to optimize the parameters of BP neural network in order to accelerate convergence and improve accuracy. The model is:
• Step 1: Initialize the network weight and bias, give each network connection weight a small random number, and each neuron with a bias will also be initialized to a random number.
• Step 2: Forward propagation. Input a training sample, and then calculate the output of each neuron. The calculation method of each neuron is the same, which is obtained by the linear combination of its inputs.
• Step 3: The gradient descent method is used to calculate the error and carry out back propagation. The weight gradient of each layer is equal to the input of the connection of the previous layer multiplied by the weight of the layer and the reverse output of the connection of the next layer.
• Step 4: The weight gradient in the third step is used to adjust the network weight and neural network bias.
• Step 5: Back propagation, Adam algorithm is used to accelerate the weight adjustment, initialize the moment vector and exponential weighted infinite norm to 0, update the parameters through vector operation, and iterate in t time from step size to 1. Sort errors and return.
• Step 6: At the end of judgment, for each sample, judge if the error is less than the threshold set by us or has reached the number of iterations. We’ll finish training, otherwise, return step 2.
Data Description and Preprocessing
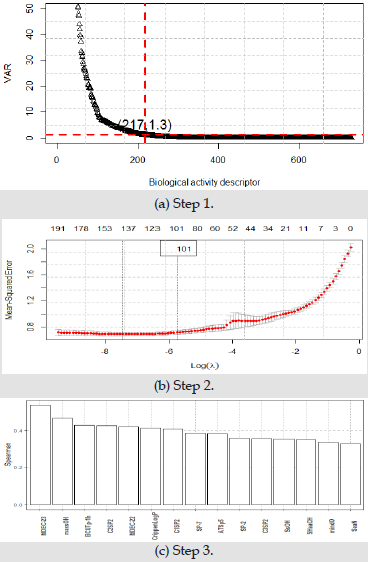
In this paper, the bioactivity description data set is used to verify the ERα activity and ADMET properties respectively. The description dataset contains 729 biological activity descriptors of 1974 compounds. Because the data dimension is too large and contains a large number of repetitions and useless variables, this paper selects 15 most representative biological activity descriptors from the 729 biological activity descriptors of 1974 compounds. Firstly, low variance filtering is used to delete the biological activity descriptors with low information, then considering the correlation and independence between variables, Lasso regression is used to select these variables, and finally considering the coupling degree between variables and ERα activity. The final 15 most representative biological activity descriptors are obtained. The specific steps are as follows:
• Step 1: Because the variance of variable can reflect the degree of dispersion, the variable with small variance contains little information, which cannot provide key and useful information for the construction of the model. Therefore, for 729 biological activity descriptors of 1974 compounds, the variance of 729 variables is calculated and arranged from large to small.
• Step 2: After cleaning the biological activity descriptors with low information or no information, use the remaining molecular descriptors to further process the repeated information of the data, so as to make the data relatively independent. In this paper, Lasso feature selection method is used to propose a variable from two variables with strong correlation to eliminate duplicate information. The essence of lasso feature selection method is to seek the sparse expression of the model and compress the coefficients of some features to 0, so as to achieve the purpose of feature selection. The parameter estimation of lasso feature selection method is as follows:
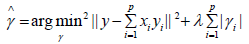
λ is a nonnegative regular parameter, which represents the complexity of the model. The greater its value, the greater the penalty of the linear model, λ Determined by cross validation.
• Step 3: Spearman rank correlation coefficient is a nonparametric index to measure the dependence of two variables, which can reflect the coupling degree between variables. This paper uses Spearman rank correlation coefficient to obtain the final 15 representative biological activity descriptors.
Three screening processes by Figure 3 shows, in step 1, 217 biological activity descriptors with variance greater than 1.3 were left. In step 2, 101 bioactivity descriptors were retained by lasso feature selection. In step 3, 101 biological activity descriptors are sorted according to Spearman rank correlation coefficient, leaving the most representative 15 biological activity descriptors. The final screening results are shown in Table 1. ADMET properties are composed of five aspects: absorption, distribution, metabolism, excretion and toxicity. The corresponding values are provided in the form of two classifications, ‘1’ represents good or yes, and ‘0’ represents poor or no. Comparison table of ADMET properties are shown in Table 2.
Model Training and Prediction
In order to avoid over fitting and improve the generalization ability of the model [10], we cut the remaining 15 bioactivity descriptors into 80% of the training set and 20% of the test set. Considering the coupling and the nonlinear relationship between the data, the neural network is used for training and prediction, the training set is used to set the model parameters, and the test set is used to calculate the default accuracy and verify the rationality of the model. When training the model, we should also consider the convergence speed of the model. Neural network is a complex structure with large amount of calculation. When there are too many input variables in the input layer and the amount of data is too large, gradient optimization algorithm is usually used to accelerate the convergence speed of neural network. Adam algorithm is used for model optimization in this paper. The results are as follows:
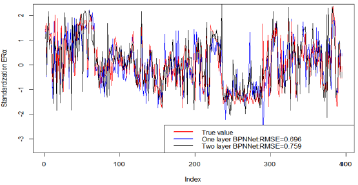
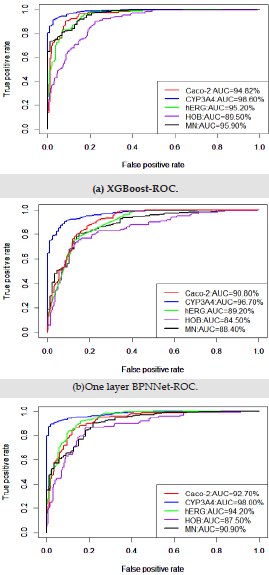
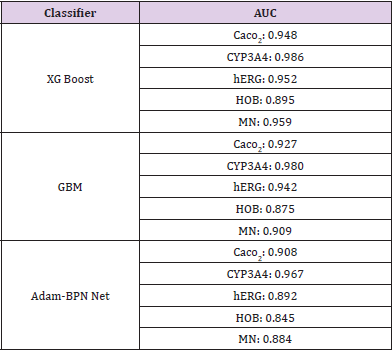
As can be seen from Figure 4, The red line is the logarithm of ERα, the blue line is the regression prediction result of neural network with one hidden layer, and the black line is the regression prediction result of neural network with two hidden layers. Among them, when the hidden layer is 1, the mean square error of prediction is 0.696, and when the hidden layer is 2, the mean square error of prediction is 0.759.Obviously, when the hidden layer is 1, the regression prediction result is more accurate, and the good prediction accuracy shows that the ERα activity can be controlled by controlling the 15 biological activity descriptors selected in this paper, so that we can inhibit the ERα activity. In order to ensure that the selected bioactivity descriptors have good medical properties, the ADMET properties of these 15 bioactivity descriptors were verified. The commonly used machine learning methods are used for multiple prediction to eliminate contingency. ROC curve shown in Figure 5. It can be seen from Table 3 that the three models show very high prediction accuracy, among which xgboost performs best. The three models show that CYP3A4 is highly coupled with 15 biological activity descriptors, HOB is the lowest coupled with one biological activity descriptor, but the prediction accuracy also reaches 0.895. This shows that the 15 biological activity descriptors selected in this paper can not only reflect ERα activity to a great extent, It can also reflect good ADMET properties.
Conclusion
The results show that the 15 biological activity descriptors selected in this paper can predict ERα activity with a low mean square error of 0.676, which indicates that there is a high coupling between them. In addition, they can also reflect the properties of ADMET at an average level of 0.948, so they have good medical value. The development of anti-breast cancer drugs is a complex and long process. In this process, it is necessary to test the effects of drugs containing various biological components on target cells. If all the combined drugs are tested, it will be a long process. In order to improve the development cycle and cost of anti-breast cancer drugs, we can consider using these bioactive descriptors to synthesize breast cancer resistant compounds. Because the experimental data are limited, the influence of these 15 bioactive descriptors on the activity of other target cells is not considered. Therefore, the bioactive descriptors selected in this paper have limitations in the effect of breast cancer. Furthermore, lasso feature selection method is used to screen bioactivity descriptors, which may omit some important bioactivity descriptors. When the synthetic breast cancer drugs are synthesized, the best value or range of bioactive descriptors can further reduce the development cost and development cycle of anti-breast cancer drugs. Therefore, in this paper, we can further study the best values of various bioactive descriptors. At the same time, we also hope that the variable screening method and validation method can be applied to more biopharmaceutical processes.




No comments:
Post a Comment
Note: Only a member of this blog may post a comment.