Fast and Presumptive Diagnosis of SARS-CoV-2 Omicron Variants vs Delta
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARSCoV- 2) Omicron variant (Lineage B.1.1.529) classified as Variants of concern (VOCs) on November 26, 2021 from World Health Organization (WHO) worldwide raises more rapidly than Alpha (Lineage B.1.1.7) and Delta (Lineages B.1.617.2 and AY various) VOCs [1,2]. The first Omicron case was sequenced at Botswana (South Africa) on November 11, 2021 and in Europe, Omicron appears firstly on 26 November, 2021 at Belgium in a young traveler woman who had been in Egypt. One day later, Italy confirmed the first Omicron detection for an Italian man who had returned from Mozambique [3]. Omicron cases decrease Delta infections, the only SARS-CoV-2 variant circulating from September to November 2021. In Italy, Omicron incidence spread from 15-20% on December 20, 2021 to 95,8% on January 3, 2022, as well as it was observed firstly in South Africa and after worldwide [4-6]. Indeed, Omicron variant was confirmed by GISAID sequences for 99.5% of global samples collected in the February 2022, sequenced and uploaded to online platform; only 0.3% of these cases were Delta and <0.1% Alpha [6].
Rapid Surveillance of SARS-CoV-2 Variants in the Diagnostic Workflow
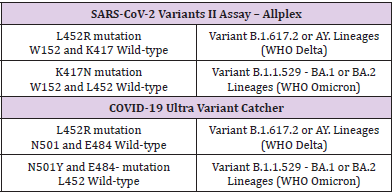
During the periodic Italian Quick Surveys on SARS-CoV-2 Variants of December 6 and 20, 2021, Liguria Reference Laboratory of Hygiene Unit - San Martino Hospital (Genoa) performed two different multiplex real-time qualitative RT-PCR assays for screening of SARS-CoV-2 variants on 100 positive nasopharyngeal swabs randomly selected among 857 new diagnosed cases (N=247 of December 6, 2021; N=610 of December 20, 2021) of Liguria (Italy). On the volume of positive SARS-CoV-2 cases notified in the previous week, sample size was established in order to reach a prevalence rate of 5% with 2% of precision. RNA was extracted by means of QIAamp Viral RNA Kits on QIAcube platform (QIAGEN Gmbh; Hilden, Germany) and processed before the sequencing with SARS-CoV-2 Variants II Assay – Allplex (Seegene Inc.; Seoul, South Korea) for detection of L452R, W152C, K417T and K417N Spike protein mutations and COVID-19 Ultra Variant Catcher (Clonit S.r.l.; Milan, Italy) which identified L452R, E484- (E484 position mutated for different amino acid variations) and N501Y mutations, according to the manufacturer’s instructions (Table 1) [7]. Detection of the SARS-Co-2 variants must be analysed by means of cycle threshold values for each specific probe, which detects the amino acid change and produces an amplification signal. Both RTPCR tests (Research use only) are validated for lower and upper respiratory tract specimens.
Table 1: Analysis and interpretation of data for Omicron and Delta variants for SARS-CoV-2 Variants II Assay – Allplex and COVID-19 Ultra Variant Catcher.
Detection of the SARS-Co-2 variants must be analysed by means of cycle threshold values for each specific probe, which detects the amino acid change and produces an amplification signal. Both RTPCR tests (Research use only) are validated for lower and upper respiratory tract specimens.
Open-Source Epidemiologic Statistics for Public Health (OpenEpi, https://www.openepi.com/) were used for a partial diagnostic evaluation of these assays calculating accuracy, sensitivity, and positive predictive values. Cohen’s κ, specificity, negative predictive values were not calculated because our real SARS-CoV-2 surveys were performed only on positive respiratory specimens and none of these were characterized by mutations of viral strains different from VOCs, Omicron or Delta. All Omicron variants (N=9, Lineage B.1.1.529.1/BA.1) with K417N, E484- and N501Y mutations and Delta variants (N=91, in particular N=5 of Lineage B.1.617.2 and N=86 of Lineages AY.) with L452R mutation were detected in advance with these assays and they were confirmed by the analysis of their consensus sequences produced through the standard reference method, next generation sequencing. Overall accuracy, sensitivity and positive predictive value of these assays were 100% (95% CI: 96.3-100%).
Discussion
High spread of Omicron emphasizes the importance of rapid epidemiological surveillance on large volume of specimens [8]. In addition, sequencing equipments and properly trained personnel are missing in some virology laboratories, especially in countries facing the pandemic crisis and the consequent economic and social crisis. This condition slows the viral variants tracking and increases their global diffusion. Indeed, a shortcoming in periodic surveillance, lack in sequencing and delay in data dissemination led to the declare the Delta emergency in India in December 2020 despite the first sequence of Delta from Madhya Pradesh has September 2020 as sampling date [9]. Molecular assays on widespread variants’ single mutations should allow epidemiological monitoring everywhere. A rapid detection of the first four Omicron cases diagnosed in Germany was reported using similar commercial kits, as well as SARS-CoV-2 Variants II Assay – Allplex and VirSNiP kits (TIB MOLBIOL, Berlin, Germany), comparing their mutations to those detected by the nanopore-based full genome sequencing with proper results [10]. To date, the diagnostic use of COVID-19 Ultra Variant Catcher for the identification of Omicron had not been reported.
Furthermore, some studies designed different home-made variants genotyping assays using specific primers and probes to detect specified mutations of SARS-CoV-2 VOCs (Alpha, Beta, Gamma, Delta and Omicron) and wild-type (non-VOCs) strains. In this pandemic period, Alpha, Delta and Omicron are only VOCs circulating; no detection of Beta and Gamma was reported from January 2022 [3]. The development of a new home-made molecular assay requires time for the validation of their diagnostic performances and a specific technical expertise [11,12]. Therefore, these home-made procedures are very useful but their applicability in some laboratories is difficult; in particular, we recommend the use of validated and available for purchase molecular assays that detect the known mutations of new VOCs for a rapid epidemiological surveillance of SARS-CoV-2 everywhere. At least, during Omicron discover and diffusion, WHO on November 22, 2021 includes in the Variants under monitoring another new variant that is B.1.640.2 with probably Cameroonian origin and harboring both substitutions N501Y and E484K. These mutations can be rapidly differentiated from Delta and Omicron variants using these same molecular SARS-CoV-2 variants assays because Spike gene of B.1.640.2 hasn’t L452R typical of Delta and K417N that it is observed among Omicron substitutions [1,13].
Conclusion
Although these assays were developed during the second and third wave of COVID-19 pandemic, between December, 2020 and June, 2021, respectively characterized by Alpha and Delta VOCs dissemination, they result very useful tools one year later to tackle the new circulating VOCs, thanks to their high accuracy, low cost and short turn-around time [5].
For more Articles on: https://biomedres01.blogspot.com/



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.