Insights of Rv2921c (Ftsy) Gene of Mycobacterium Tuberculosis H37rv To Prove Its Significance by Computational Approach
Introduction
Tuberculosis is a fatal disease which is broadly conveyed and open. It is caused by Mycobacterium tuberculosis H37Rv (M. tuberculosis H37Rv) which is a gram-positive and aerobic bacterium [1]. Pathogenic strain H37Rv of this bacterium is differing from other non-virulent strains like Mycobacterium smegmatis (M. smegmatis) in various means [2]. After crossing the respiratory tract, this bacterium permanently resides in the alveolar macrophages where they reside for long time without any hindrance by host immune system [3]. It is very important to decrease the level of this disaster which appears to be at its peak and to comprehend this situation, struggle is going on. As it is already known that, BCG is the only candidate vaccine available till now for the curative therapy of tuberculosis (TB). There are many more drugs used in the treatment of this disease but not as effective as BCG vaccine, although in some of the cases BCG also fails to protect an individual from the drastic nature of this disease [4]. In the year 2016 only, there were 6 million novel cases had been reported which develop resistance to Rifampicin (RRTB)–the most effective and first line drug to battle TB [5]. Although there were some strategies made by WHO to end this disease and the management also succeed to a minor level, we right now deal with the situation that need a quick treatment with respect to the total population and the environment of our nation. M. tuberculosis H37Rv and HIV co infection are prone for diminishing the CD4+ T cell population which is known as cell mediated arm of immunity and this co-infection is now widespread in all over the world [6]. Persons with this co-infection are lessening host cell survival due to multifaceted nature of both these bacterium [7]. Thus, as to figure out problem of the pathogenesis of this bacterium, we have to figure out important aspects of this bacterium that may play significant role in its survival inside host cell and avoid many host immunological barrier [8]. To grasp trouble pathogenesis, it is imperative to portray significant highlights of M. tuberculosis H37Rv that sanction it to evade the host barrier framework and add to its destructiveness [9]. Many previous studies show the importance of GTP binding and hydrolyzing genes in the survival of the many prokaryotes as well as eukaryotes.
Methods and Material
Retrieval of Protein Sequence Database
Mycobrowser database has been used for retrieving sequence (gene and protein) of ftsY (Rv2921c) gene [29]. This ftsY (Rv2921c) protein sequence has been disentangled in FASTA format ftsY is predicted to be involved in insertion and reception of various proteins in the outer side of cell membrane and contains ATP/GTP binding motif.
For the analysis of multiple sequences alignment of ftsY gene (Rv2921c) of M. tuberculosis had been done with ftsY gene of Mycobacterium bovis (M. bovis), Mycobacterium marinum (M. marinum), M. smegmatis and Mycobacterium leprae (M. leprae). MUSCLE online server for multiple sequence alignment approach for ftsY protein analysis. MUSCLE remains for Multiple Sequence Comparison by Log-Expectation it is professed to accomplish both better normal exactness and preferred speed over ClustalW2 or T-Coffee, contingent upon the picked choices [30,31].
STRING database server is utilized for demonstrating the protein-protein association between two or more entities. In the cell cytoplasm, a protein may collaborate with different proteins and work in the web-like manner. The associations incorporate direct (physical) and aberrant (functional) interactions; They branch from computational expectation, from arrangement pass on among life forms and from connections collected from other primary databases [32]. The total number of interactions for each dataset had been kept in STRING and measure somewhere in the range of 0 and 1. The principle of its working is as the score is < 0.4 means low interaction, score is between 0.4 to 0.7 means medium interaction and score is >0.7 is high interaction [33].
Protein Subcellular Localization and prediction confined protein destinations. TBpred is a subcellular localization prediction method for mycobacterial proteins depend on support vector machine learning (profile portion SVM) to foresee the local subcellular sites. Several parameters may be tuned for their appropriate values to get optimum results. The nth SVM model learns from nth class samples with positive labels and rest other samples with negative labels. Prediction of an unknown sample is based upon the maximum score out of four scores, generated by four models specific to four different subcellular compartments [34].
DEPP (Disorder Enhanced Phosphorylation Predictor) server evaluates the number of phosphorylated serine, threonine and tyrosine site. This server is depending on the Support Vector Machines prepared on succession profiles improved by data from the spatial setting of tentatively distinguished P-locales. DEPP server predicted the phosphorylated sites of serine, threonine and tyrosine with accurate score. DEPP server is able for predicting phosphorylation by the serine kinases PKA, PKC, MAPK, CKII and by the tyrosine kinases SRC. The nature of expectations is incredibly reliant on the nature of submitted protein structures. Incorrect or inadequate protein structures may prompt wrong forecasts [35,36].
The prediction of (B & T-cell) epitopes found in ftsY (Rv2921c) protein was done by various online bioinformatics tools. B-cells epitopes prediction was done by using BCPREDS server, T-cell epitopes prediction by the ABCpred server and (MHC-Class II Binding Peptide Prediction) is done by ProPred tool [37-40].
Structure modeling of ftsY protein was done by using Iterative Threading Assembly Refinement (I-TASSER). I-TASSER is utilized for the protein structure and function expectation [41-44]. I-TASSER needs the FASTA prearranged sequence of protein and assembles the 3D model of protein by Ab Initio display approach. I-TASSER server is an online stage for protein structure and functions forecasts [45]. I-TASSER pursues three phases to envision the 3D model of the protein. For advance illustration of the secondary structure of the protein, this tool secretly introduced Local Meta Threading Server (LOMETS) which uses H, E and C articulate for alpha-helix, beta-sheet and curl respectively. In I-TASSER server there are also predicted the desired dynamic binding sites of our selected protein were anticipated by COACH online server. Prior to dynamic site-specific docking, the affirmation of actual binding pocket is important. The binding pocket is the site of protein where the ligand interacts reversibly or irreversibly [46]. COACH server is a metaserver, it starts from given structure of target protein; At that point it will make correlative ligand restricting site forecast using the two relative systems, TM-site and S-site, which perceive ligand restricting plan from the database (BioLiP) protein work database by restricting specific substructure and collection profile examination. In the COACH server, yield has positioned top 10 displays by the bunch measure, given C-score, PDB hit, ligand name, complex structure download and agreement restricting buildup. Range estimations of C-score prediction lie somewhere in the range of 0 and 1, where the most noteworthy score demonstrate greater unwavering quality [47].
The evaluation of approval of the created protein structure has been completed by online server RAMPAGE (Ramachandran Plot Analysis). The RAMPAGE server endorses the protein structure on the hypothesis of φ, ψ purpose of individual stores [48-50]. The approval of protein was performed by the structure Analysis and confirmation server rendition for (SAVES) or, in other words server that depend on checking the stereo compound nature of a protein structure by breaking down residue geometry and by and large structure geometry [51,52].
In the Mutational assessment of ftsY (Rv2921c) had been finished by via I-MUTANT 3.0 suite server. For studying of all the proteins, thinking about analysis of protein quality, free vitality change (ΔΔG) upon single point transformations may in engage the clearing up of process. The vast majority of the ΔΔG esteem is about zero (around 32% of the ΔΔG edifying record ranges -0.5 kcal/ mole) and both the consideration and suggestion of ΔΔG might be either positive or negative for a similar change blurring the relationship among imprecise and expected ΔΔG esteem. Remembering the last goal to vanquish this issue, we describe another pointer that confines among the three change classes: destabilizing changes (ΔΔG<−1.0 kcal/mol), counter balancing changes (ΔΔG>1.0 kcal/mole), and impartial changes (−1.0≤ΔΔG≤1.0 kcal/mole). For the I-MUTANT 3.0 suite score, DDG<-0.5 means (extensive decline of stability), DDG>0.5 infers (increment of stability) and - 0.5<=DDG<=0.5 means (impartial stability) [53,54]. For the figure of protein, consistency change upon a singular point alteration was foreseen by I-MUTANT 3.0 Suite server.
Docking has been studied for confirming protein-protein and protein-DNA interaction analysis. Complexes formed after docking were visualized by the PyMOL. Molecular docking of ffh (Rv2916c) with wild type and mutant ftsY protein revealed the variation of binding energies, formation of hydrogen bonds and their distances. Docking of the two proteins we have been used HDOCK (http:// hdock.phys.hust.edu.cn/) docking server [55]. This server takes input in form of either FASTA sequence or PDB structure. This feature of taken up FASTA sequence makes this server easily available for new comers. The output of the server contains docking score and RMSD value. The lower the RMSD value stronger is the association. HDOCK predicted the protein–protein and protein– DNA/RNA docking. The interactions of the protein-protein and protein–DNA/RNA play an essential role in the assortment of biological process. For the docking, HDOCK is the novel tool for hybrid docking algorithm of template-based modeling and free docking, in which cases with the misleading templates it can be rescued by the free docking protocol. The docking process is fast and consumes about 10-20 min for a docking run [56,57]. HDOCK server performance will become enhanced when more predictions were considered.
Result
Retrieval of Target Protein Sequence
The genomic and proteomic sequences for the gene ftsY (Rv2921c) have been retrieved in FASTA format from Mycobrowser database. The gene is 1269bp long and encodes protein of 43kDa. The predicted function of this protein is involving in reception and insertion of the protein at membrane site. Probably it functions as Signal Recognition Particle (SRP). The protein sequence contains four GTP binding motif (DXXG).
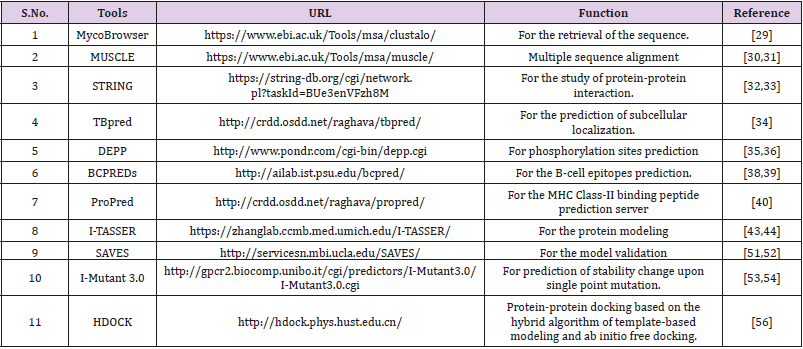
For the prediction of protein-protein interaction of ftsY protein is involved with the secretion system, involved the insertion of promising membrane proteins and it is in cytoplasmic membrane. The ftsY protein acts as a receptor for the complex formed by the signal recognition particle and the ribosome-nascent chain. The prediction of ftsY functional partner by STRING are the ffh, rplJ, rplQ, Rv3362c, rplK, rplR, rplS, rpmA, rplW, rpmG1 proteins due to the decreasing order of the String server version 10.5 score. The interactions score of ftsY and ffh interaction are very high 0.995 and both gene having same function and act as SRP protein Figure 1.
 Multiple Sequence Alignment
Multiple Sequence AlignmentMultiple sequence alignment of the protein ftsY (Rv2921c) of M. tuberculosis with other species of the bacterium like Mycocaterium bovis, Mycocaterium marinum, Mycocaterium smegmatis, Mycocaterium leprae has been done by using MUSCLE online server. After putting sequence in FASTA format for all five proteins the server aligns sequence and gives the perfect matches. The percent identity matrix proves its identity among all species. The result also shows the presence of consensus sequence DXXG in all sequences (Figure 2). The presence of the motif in all species is might be an indicator for its importance of the process in the survival of these prokaryotes.
 Prediction of Sub Cellular Localization
Prediction of Sub Cellular LocalizationPrediction of the subcellular localization of a protein by using the TBpred server the length of this gene is 422 amino acid residues and the selected approach are Dipeptide composition based SVM. There are different class wise scores from SVM models like cytoplasmic protein, integral membrane protein, secretory protein and protein attached to membrane by lipid anchor. At last, this TBpred server finally predicts that ftsY protein is localized in membrane as an integral membrane protein.
 Phosphorylation Site Prediction
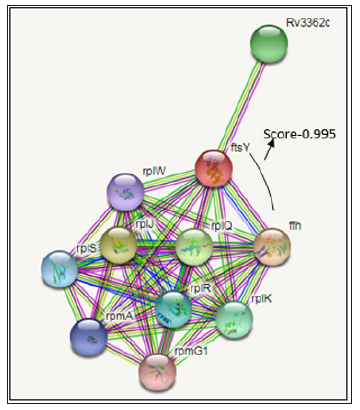
Phosphorylation Site PredictionDEPP (Disorder Enhanced Phosphorylation Predictor) server evaluates the number of phosphorylated serine, threonine and tyrosine site. Notwithstanding serine, threonine and tyrosine result shows that in ftsY protein there were 8 phosphorylated serine residues out of 18 residues, 9 phosphorylated threonine residues out of 25 residue and 1 phosphorylated tyrosine residue out of 2 residues. In DEPP statistic score are 44.44% are phosphorylated serine, 36% are phosphorylated threonine and 50% tyrosine, phosphorylation prediction also shown in Figure 3. The nature of expectations is incredibly reliant on the nature of submitted protein structures. Incorrect or inadequate protein structures may prompt wrong predictions.
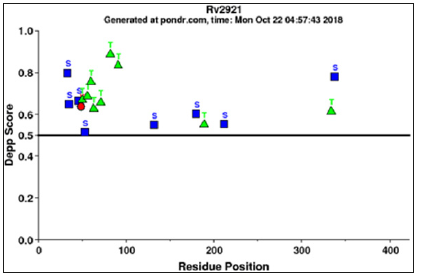
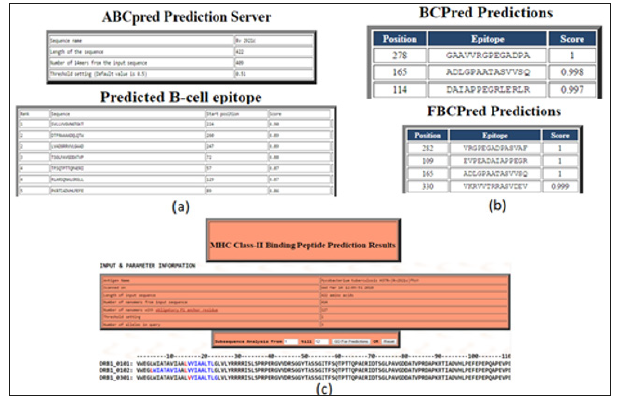
The prediction of B-cell epitopes by ABCpred server which has predicted the epitopes on this gene taking the overlapping window of 14 amino acids that consequences in the preeminent possibility of the score is 0.90 from the residues region “SVLLVVGVNGTGKT” that starts at 224th position as shown in supplementary (Figure 4a). The BCPREDS server are B-cell epitopes prediction which have shows two types prediction as Fixed length epitopes prediction by BCPred and flexible length epitopes prediction by FBCpred which have been shown in (Figure 4b) with the set specificity at 90% and epitopes length set on 14. B cell epitopes prediction fixed length method are predicted on residues 278th -291th, 165th-178th, and 114th-127th and for Flexible length epitopes prediction residues committed are as 282th -295th, 109th -122nd, 165th-178th and 330th- 343th in the ftsY protein. For the T-Cell epitopes prediction there is multiple DR-β1 (DRB) alleles were used like HLA-DRB1*0101, HLA-DRB1*0102, HLA-DRB1*0301 which is the T-cell epitopes prediction for the prediction with the MHC Class-II binding region in the antigenic protein sequence of ftsY protein. The predicted binder was visualizing in graphical peak interface as well as in color residue in an HTML interface. Two consensus epitopes were LVVIAALTL in (DRB1_0101) at position 15th-23rd in (DRB1_0102) epitopes are present were – LWIATAVIA and LVVIAALTL, at the 5th -13th and 15th-23rd and in (DRB1_0301) VVIAALTLG residue are present 16th-24th position. In MHC Class-II binding peptide prediction result there are individual alleles at 1% threshold as shown in T-cell epitopes prediction. While at 3% threshold it gives another residue, which is not mention in this research. For T-cell epitopes prediction result are shown in Figure 4c.
 Model Building
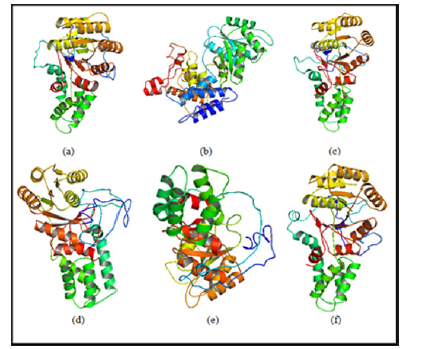
Model BuildingThe structural modeling of the wild type and mutant ftsY (Rv2921c) protein and its interactive partner ffh (Rv2916c) protein were prepared from I-TASSER. The quality of modelled protein depends upon the percentage of the favorable region lies above 90% of the value of C-score and RMSD value. The C-score is the confidence score for each model. It is computed by threading layouts arrangement. The C-score changes inside the range from -5 to 2 and higher certainty show is controlled by the higher estimation of C-score. At long last, I-TASSER creates top 5 models according to C score and positioned by group measure among which the figure with higher C score. For protein modeling we modeled the ffh (Rv2916c), ftsY (Rv2921c), mutant ftsY (D45) in this protein modeling we mutate 45 number residue aspartates into alanine, mutant ftsY (D71) residue, mutant ftsY (D312) residue and mutant ftsY (D367) residues as shown in Figure 5.
 Model Validation
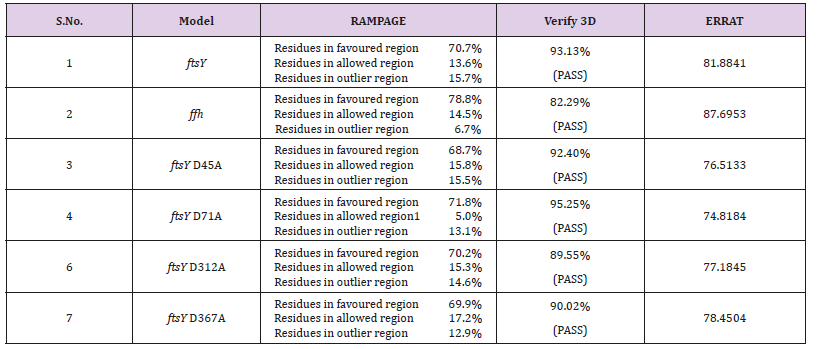
Model ValidationAfter modelling of structure, the protein structure was validated through and SAVES server (RAMPAGE, ERRAT and Verify3D). The demonstrated protein was validated by RAMPAGE (Ramachandran plot investigation) which is an online server. After examination of Ramachandran plot of our proteins, the structure demonstrated that of have been present in a favored region. Although, other residues were laid in the allowed region and number of residues were laid in outlier region. These parameters of protein structure demonstrating that our displayed protein was of good quality stable and adequate. ERRAT is an online server which approves the protein structure on the premise of the nuclear connection between various sorts of atoms. The ERRAT analysis overall quality factor of our model protein is good and satisfactory. The Verify3D strategy evaluates protein structure by utilizing three-dimensional profiles. This program examines the similarity of a nuclear model (3D) with its own amino acid sequence which is 1 dimensional. Every deposit is doled out a basic class in radiance of its area and condition (alpha, beta, circle, polar, non-polar and so on). The score ranges from -1 (poor score) to +1 (great score). 82.29 - 95.25% of the buildup had a found the middle value of 3D-1D score >=0.2 that is perceptive for our demonstrated protein result. is pass as shown in Table 2 Verify 3D result. The protein model ftsY (Rv2921c) validation which is done by SAVES metaserver results convoluted that all of the protein model build are good and satisfactory which is shown in Table 2.
 Mutational Analysis
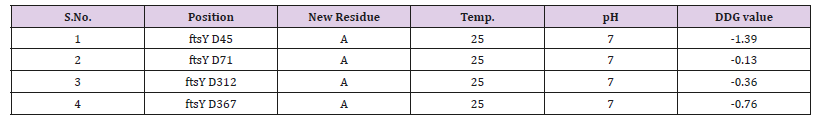
Mutational AnalysisIn ftsY (Rv2921c) protein had been characterized as a GTP binding protein which has consensus binding sequence DXXG which promote the link sub sites for binding in Mg2+ and -phosphate of GTP. In the consensus sequence DXXG, aspartate is crucial for GTP binding and hydrolyzing activity; Therefore we study the prediction of stability change upon the single point protein mutation by I-Mutant suite. Earlier studies have been shown that for GTP binding protein, when aspartate mutate with the alanine then the functional protein may loss the function. So, we mutate the DXXG aspartate into the Alanine for predicting the stability of the protein after change the single amino acid. In I-Mutant suite studies, we have seen that there are four consensus sequences in this protein on residue 45, 71, 312 and 367. After the analysis of predicted score by I-Mutant we have found that large stability decreased on the 45-position residue because the parameter of stability decreasing value is when -0.5 or below value towards negative and our outcome result is -1.39 which clearly shows at the position on 45 there will largely decreasing value are shown in Table 3.
 Molecular Docking
Molecular DockingIn the docking content, we here doing protein-protein docking for knowing the interaction changes upon mutation of important residue aspartate involved in GTP binding and hydrolyzing motif in the protein. Aspartate would be mutating with alanine. The wild type ffh protein dock with wild type ftsY as a control value for our study for the experimental study like wild type ffh protein dock with mutant ftsY (D45A), ftsY (D71A) , ftsY (D312A) , ftsY (D367A) to identify change in binding patterns using HDOCK docking server [14,15] as shown in Figure 6. The binding energy and formation of hydrogen bonds to each molecule by the drug were calculated and the RMSD value was 0.38 the highest for D312 are seen in Table 4.
 Table 4: Molecular docking by HDock server.
Table 4: Molecular docking by HDock server.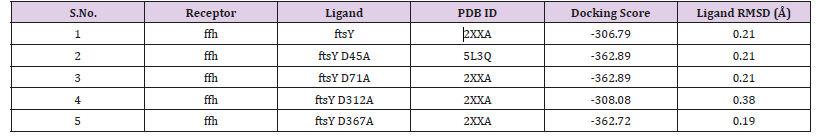
Discussion
In the current circumstance, we can see that there no defensive and healing treatment to destroy tuberculosis totally aside from BCG. Past many years of research as of now demonstrates that BCG gives constrained insurance against tuberculosis yet bombs in securing MDR, TDR and XDR instances of tuberculosis. There is a persistent exertion has been put by researchers with the end goal to build the adequacy of the antibody and in looking for new medication targets. GTP binding genes come out to be as novel targets for treatment of this disease [58-59]. GTP binding and hydrolyzing protein ftsY (Rv2921c) has been proved for possessing the same activity in many studies, this gene strongly interacts with ffh gene (Rv2916c) with score of 0.995 [32]. At the other hand, ftsY protein is also interacts with another predicted partner Rv3362 which also possess GTP binding activity [33]. Multiple sequence alignment result shows that this gene is universally present in all species of this bacterium with having some minute differences. All species contains same GTP binding motif DXXG at same location [30,31]. This protein is predicted to be present as an integral membrane protein [34]. Phosphorylation site prediction gives us result that this protein phosphorylated at serine, threonine and tyrosine [35,36]. The prediction of (B and T cell epitopes prediction) has been done by using ABCpred, BCpred, FBCPred and ProPred server [38-40]. 3D model of this protein has been made by I-TASSER server [43,44] and validated by SAVES server [51,52]. Mutational analysis has been done by I-Mutant 3.0 server and it shows that mutation on 45 residue aspartates with alanine at 25°C temperature and pH 7 noted the larger decrease in stability [53,54]. Molecular docking results of these proteins before and after mutation prove that interaction between these two proteins decrease maximal at position 312 residue with having value of 0.38 [56]. In summarizing our work we can narrate the essentiality of ftsY gene in secretion process and thus in pathogenesis of this bacterium. The two above mentioned proteins work in cooperative manner and produce a summative effect. The two proteins is key regular for the co-translational secretion system and thus might also play an important role in pathogenesis of the bacterium.
Conclusion
We need an emergent step to stop TB epidemic all around the world. This study concludes with knowledge of some important aspects of ftsY gene and its interaction with ffh gene. This interaction is very much important for one of the secretory pathways assembled in prokaryotes. Although the experiments enlisted in this manuscript are not enough, but these experiments provide us with better knowledge and initial steps in further in vitro and in vivo experimental works.
More BJSTR Articles : https://biomedres01.blogspot.com



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.