Introduction
Milk is the most popular food for human consumption and considered as a complete and nutritious food; not only for the new-born but for all age groups in both rural and urban people all over the world [1]. It is the major source of regular income for dairy producers because it is produced and sold daily Dugdill et al. However, it serves as an excellent growth medium for a wide range of microorganisms [1]. In Ethiopia, urban and peri-urban dairy farming creates an important sector of the agricultural production system [2]. Propensity of fast increasing human population together with growing urbanization creates increased demand for milk and milk products. Post-harvest losses of about 40% milk and its products have been reported from milking to consumption. Such losses are mainly attributed to mishandling in dairy value chain from farm to consumers. These include, contamination during milking, prolonged storage, deliberate adulteration, substantial handling, transportation and distribution system [3].
The natural raw milk obtained from the mammary gland of healthy animal is usually with low microbial load and the application of all hygienic measures during milking prevents milk from contaminating as well. The bacteria can access to the milk through colonization of the teat canal or an infected udder (clinical and subclinical mastitis) or gets contaminated from milk utensils or water supply used. The presence of bacteria in milk has many undesirable effects on the quality and safety of milk and its products. Milk contaminated by high levels of bacteria usually becomes unsuitable for further processing [4]. Assessment of bacterial load is a frequently used procedure to measure the quality of milk. The Standard plate count (SPC) and coliform plate count (CPC) methods are universal methods to estimate the total aerobic and coliform bacterial numbers present in raw milk [5]. E. coli is a subgroup of the fecal coliform group. Although most E. coli bacteria are harmless, they are among many pathogenic microorganisms which can access to milk and other dairy products, and considered as a reliable indicator of contamination by manure, soil and contaminated water [6].
E. coli is a normal inhabitant of the intestines of animals and humans but its recovery from food may be of public health concern due to the possible presence of enteropathogenic and toxigenic strains which lead to sever gastrointestinal disturbance [7]. However, the other toxigenic strains like E. coli O157:H7 cause life threating syndromes [8]. E. coli O157:H7 causes the majority and most severe outbreaks of gastrointestinal illnesses that range from asymptomatic conditions to mild bloody diarrhea with severe abdominal pain. Occasionally other complications such as hemorrhagic colitis, hemolytic-uremic syndrome and thrombotic thrombocytopenic purpura (TTP) may occur [9]. The treatment of illnesses caused by E. coli O157:H7 often requires antimicrobial therapy. The repeated and unsuitable use of antibiotics has led to an increasing rate of antimicrobial resistance [10].
There is worldwide concern about the appearance and rise of bacterial resistance to commonly used antibiotics. The evolution of increasing prevalence and dissemination of pathogenic bacteria resistant to multiple antimicrobial agents is currently recognized as one of the most important problems in global public health [11]. The rapid spread of antibiotic resistance genes, facilitated by mobile genetic elements such as plasmids and transposons, has led to the emergence of multidrug resistant strains of many clinically important species that nowadays frequently leave clinicians out of therapeutic options [12]. The aim of thriving a number of dairy farms is to maximizing production and profitability of the diary sector. However, quality control is not habitually employed at individual farm level, and there is shortage of data pertaining to the level of spoilage organisms and antibiotic resistant pathogens in commercially available milk along the dairy value chains in country. Thus this study was designed with the following objectives:
a. To assess handling practices and determine bacteriological quality of milk along the value chain from farm to fork
b. To determine to what extent the dairy farms and milk retailers serve as sources of E. coli and E.coli O157: H7
c. To identify the antimicrobial susceptibility patterns of E.coli O157: H7
Materials and Methods
Description of the Study Area
The study was conducted from January 2016 to April 2017 in Addis Ababa. Addis Ababa is the capital city of the Federal Democratic Republic of Ethiopia, which is divided into ten subcities namely Addis Ketema, Akaky Kaliti, Arada, Bole, Gullele, Kirkos, Kolfe-Keranio, Lideta, Nifas Silk-Lafto and Yeka [13]. Addis Ababa is situated at latitude of 8o55’ and 9°3’North and 38°43’ and 38o50’East, and it has an area of 51,000 hectare in the central highlands with an average altitude of 2000-2560 meters above sea level. The area is characterized by a relative humidity varying from 70% to 80% during the rainy season and 40% to 50% during the dry season, and bimodal rainfall with an average of 1800mm, the highest percentage of rain falls is during the long rainy season from June to September. The short rainy season is from February to April. Its annual average maximum and minimum temperature are 26 °C and 11° C respectively; with an overall average of 18.7 °C [14]. The city has about 5, 200 dairy farms with a total of 38,572 dairy cows [3].
Study Design and Population
A cross sectional study was conducted from January 2016 to April 2017 in four selected subcities of Addis Ababa. The study involved different actors along the dairy value chains; farmers, milk vendors and restaurants from each sub cities. Two types of dairy farms, small and medium scale dairy farms, with their lactating cows were involved in the study. The inclusion criteria of the study were based on the availability of milk during the time of sample collection, willingness to participate and the accessibility of different actors in the sub cities.
Selection of Study Districts and Dairy Farms
From ten subcities, four subcities (Akaky Kaliti, Nifas silk- Lafto, Bole and Yeka) were purposively selected for data and sample collection. Prior to data and sample collection, all the farmers who had small and medium scale farms in each selected sub cities were identified and each listed on a piece of paper. A total of 70 small and medium scale dairy farms were randomly selected and milk and swab samples were taken and questionnaires designed to focus on pre and post-harvest milk handling practices were administered based on their willingness.
Milk Vendors and Restaurants Selection
Prior to sampling, milk vendors and restaurants from each subcities were identified. About 32 milk vendors and 44 restaurants were selected by purposive sampling. Raw milk and swab samples from vendors, and boiled milk from restaurants were taken and questionnaire designed to focus on the type of milk sold, source of milk and hygienic practices were administered to selected vendors and restaurants which had milk during the time of sample collection.
Sample Size Determination
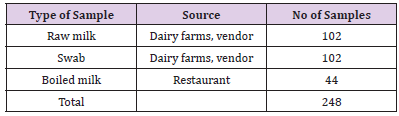
All dairy farms, vendors and restaurants fulfilling the inclusion criteria were considered for sampling. Accordingly, a total of 248 (102 raw milk, 44 boiled milk and 102 equipment swab) samples were collected from selected small and medium scale dairy farms, vendors and restaurants based on availability and accessibility of time for sampling and laboratory works.
Method of Data Collection
Questionnaire Survey: A semi structured questionnaire was used to collect data through interviews. Data obtained from respondents was focused on pre and post-harvest milk handling practices (barn type and cleaning practices, source of water used for cleaning of udder, milker and milk utensils, housing management, and awareness regarding animal management). In addition, milk vendors, restaurant workers was interviewed on the quality of milk they handle, possible sources of microbial contamination and associated problems. The questionnaires were administered through face to face conversation. While administering questionnaires, direct observation on general cleanliness and hygienic practices with regard to milk was also done and noted. Upon finishing of the administration of questionnaires, milk and swab samples were collected for laboratory analysis.
Sample Collection and Transportation: Milk and equipment swab samples were collected from the selected actors along the dairy value chain (Table 1). In that aspect, raw milk and swab samples were collected from medium and small scale dairy farms, milk vending shops, and boiled milk from restaurants. At farm and vender level, swab samples were taken from milk containers before milking and storage, and milk samples were obtained directly from the containers used during milking, distribution and storage. About 10 to 20ml of milk and proper swab samples were collected and put in sterile bottles and in 5ml of peptone water respectively, properly labelled and kept in ice box with ice packs. Within four to six hours samples were transported from the study area to microbiology laboratory of College of Veterinary Medicine and Agriculture, Addis Ababa University, Bishoftu, for microbiological analysis.
Table 1: Types of samples collected for laboratory analysis.
 Bacteriological Laboratory Analysis
Bacteriological Laboratory AnalysisBacterial Load Determination: The bacteriological tests considered for the determination of the bacterial load in raw and boiled milk samples were Total Aerobic Bacterial Count (TABC) and Total Coliform Count (TCC).
a. Total Aerobic Bacterial Count (TABC): The total aerobic bacterial count was made by adding 1 ml of milk sample into sterile test tube having 9ml peptone water. After thoroughly mixing, the sample was serially diluted up to 1:10-7 and duplicate samples (0.1ml) were pour plated using 15-20ml standard plate count agar solution and mixed thoroughly. The plated sample was allowed to solidify and then incubated at 37ºC for 24 hours [15].
b. Total Coliform Count (TCC): One millilitre of milk sample was added into sterile test tube having 9 ml peptone water. After mixing, the sample was serially diluted up to 1: 10-5 and duplicate samples (0.1ml) were pour plated using 15-20ml Violet Red Bile Agar solution (VRBA). After thoroughly mixing, the plated sample was allowed to solidify and then incubated at 37 ºC for 24 hours [15].
After incubation, all colonies including those of pin point size in SPCA medium and purplish and dark red colonies in VRBA medium were counted under colony counter and results from each SPCA plates which contained 25 to 250 colonies per plate whereas, less than 100 coliform colonies from VRBA were recorded.
After counting and recording bacterial colonies in each petri dish the number of bacteria in millilitre milk was calculated by the following formula given by [15] as referred by [16].

Where: N = number of colonies per millilitre of milk, ΣC = sum of colonies on plates counted, n1= number of plates on lower dilution counted, n2 = number of plates in next higher dilution counted and d = dilution from which the first counts are obtained.
Isolation and Identification of E. coli and E.coli O157:H7: The methods used in isolation and identification of both E.coli and E.coli O157:H7 were according to the techniques recommended by [5]. MacConkey agar media which is used as a differential media for identification of E. coli was prepared according to the manufacturer’s instructions and milk and swab sample was streaked onto agar plates. Then, the plates were inverted and incubated at 37 ºC for 24 hours. After incubation, the plates was examined for E.coli colonies. Colonies of E. coli grown on MacConkey Agar persummed to be dry, medium in size, pink in colour and grown in single or groups. The presumed colonies was again sub-cultured on selective medium Levine Eosin Methylene Blue (EMB) Agar and incubated at 37 ºC for 24 hours. Morphologically typical E. coli colonies was producing metallic sheen and sub-cultured further onto Nutrient Agar for biochemical confirmation. To differentiate E.coli from other Enterobacteriaceae biochemical tests like indole, methylene red, citrate and Voges-Proskauer (IMVC) tests were performed to wellisolated colony from nutrient agar plates to confirm the presence of E. coli in the test samples.
E.coli O157:H7 is difficult to detect with ordinary methods. Most strains of E.coli O157:H7 ferment lactose rapidly and indistinguishable from non-O157:H7 E.coli on conventional enteric media. E.coli O157:H7 does not ferment sorbitol, unlike most non- O157:H7 E.coli, salmonella, and citrobacter. The use of a selective/ differential medium such as Sorbitol MacConkey Agar has been found to facilitate detection of E.coli O157:H7. Sorbitol MacConkey Agar media was prepared according to the manufacturer’s instructions and pure colonies of E.coli were streaked onto agar plates.Then, the plates were inverted and incubated at 37ºC for 24 hours. After incubation, the plates was examined for E.coli O157:H7 colonies. Colonies of E. coli O157:H7 grown on Sorbitol MacConkey Agar presummed to be non-O157:H7 E.coli appeared pink whereas E.coli O157:H7 appeared pale or clear colourless. E.coli O157:H7 latex test was used to presumptively identify E.coli O157:H7 from sorbitol-negative colonies grown on SMAC primary culture plates. When test latex particles are mixed with fresh colonies of O157 and/or H7 strains of E.coli, an immunochemical reaction occurs, resulting in agglutination.
Antimicrobial susceptibility of E.coli O157:H7
The antimicrobial susceptibility test was performed according to the standard agar disk diffusion method using commercial antimicrobial disks. Each isolated bacterial colony from pure fresh culture was transferred in to a test tube of 5 ml Tryptone Soya Broth and incubated at 37oC for 6 hours. The turbidity of the culture broth was adjusted using sterile saline solution or added more isolated colonies to obtain turbidity usually comparable with that of 0.5 Mc- Farland standards. Mueller-Hinton agar plates were prepared according the manufacturer’s instruction. A sterile cotton swab was immersed into the suspension and rotated against the side of the tube to remove the excess fluid and then swabbed in three directions uniformly on the surface of Mueller-Hinton agar plates.
After the plates dried, antibiotic discs were placed on the inoculated plates using sterile forceps. The antibiotic discs was gently pressed onto the agar to ensure firm contact with the agar surface, and incubated at 37oC for 24 hours. After incubation the diameter of inhibition zone formed around each disc was measured using a black surface, reflected light and transparent ruler by lying it over the plates. The result was classified as sensitive, intermediate, and resistant according to the standardized table supplied by the manufacturer [17].
Data Management and Analysis
All the raw data were coded and entered in Microsoft Excel-2013 spread sheet. The total aerobic bacteria and coliform count data was transformed to log values before subjected to statistical analysis. The analysis was carried by Stata version 13.0 (2013). Mean and overall mean were summarized using descriptive statistics. Association of isolation with considered variables (sample types, sample sources and districts) was determined by Chi-square. The mean TABC and mean TCC difference between sample sources were compared by using ANOVA test. While the mean TABC and mean TCC difference between samples types were analysed by using t-test statistics. The significance level was set at 95% CI and critical p value (p < 0.05).
Results
Demographic Characteristics of the Respondents
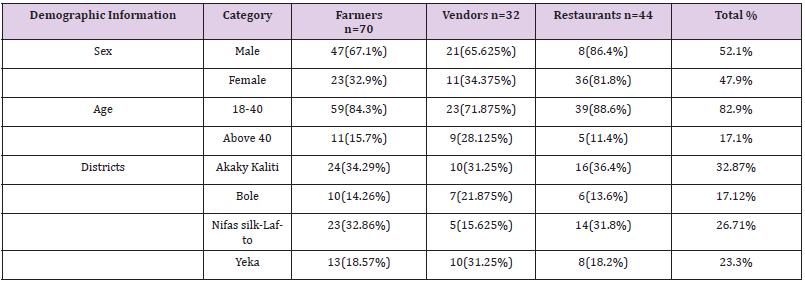
From a total of 146 (70 farmers, 32 vendors and 44 restaurants) respondents, there were more males in dairy farms and vendors compared to females but more females involved in restaurants than males (Table 2). The highest age proportion of the respondents were ranged 18-40 years which accounts about 82.9% while the rest of the respondents were above 40 years which holds 17.1% (Table 2).
Table 2: Demographic characteristics of the respondents (N=146).
 Hygienic Handling Practices
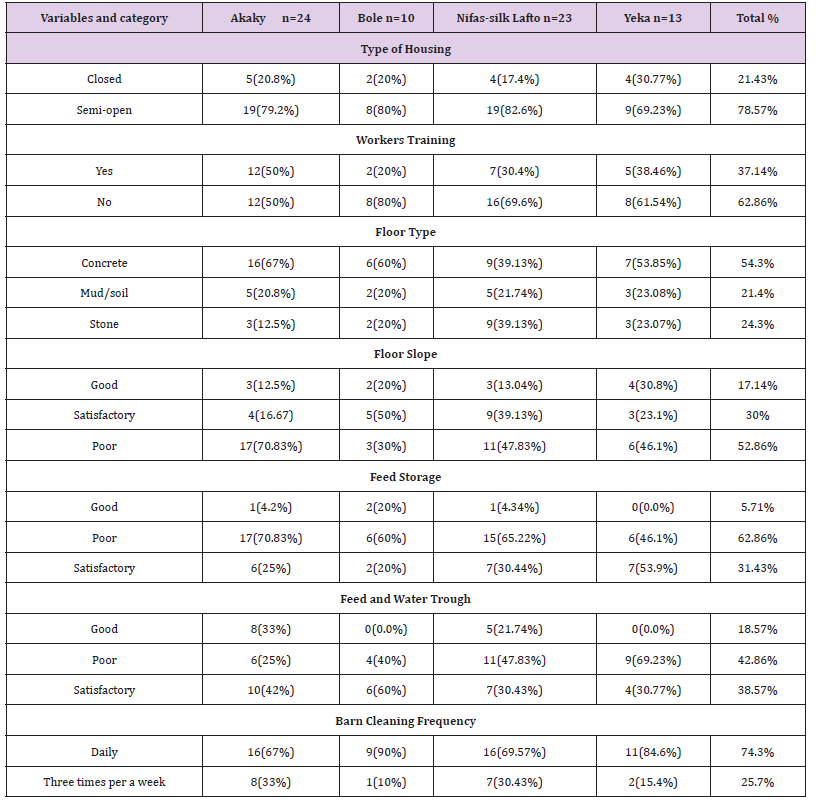
Hygienic Handling PracticesType of Housing and Cleaning Practices: About 62.86% of the farm owners and workers were not given training about farm management systems and related practices and most (78.56%) of the dairy barns were semi open and built with metallic or iron sheets even though the floor materials in most farms were generally of concrete or cemented followed by stones and mud or soil floor (Table 3). There were poor status of floor slope and feed storage area in the barn but feed and water trough was satisfactory in most dairy farms. In addition most of them were cleaned the barn once per day (Table 3).
Table 3: Type of farm housing, workers training and cleaning practices in dairy farmers (N=70).
 Hygienic Condition of Cows and Milkers
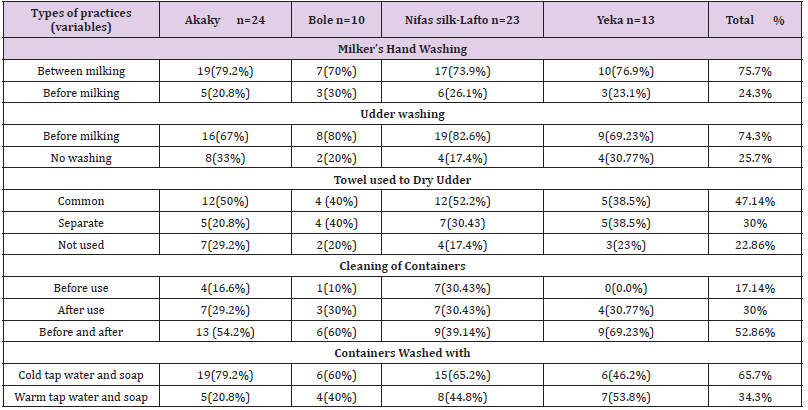
Hygienic Condition of Cows and MilkersThe main source of water used for sanitary activities was tap water and always used during milking in untreated form. During observation, the most common type of containers used during milking, storage and distribution were the wide necked plastic containers. There were no cold storage facilities as all the milk transactions from milking; storage and transportation were being done under room temperature. More than 75.7% of farmers did not clean their hands between milking and about 25.7% of them never wash udder (teats) and did not clean barns before milking (Table 4). All the farmers reported to do hand milking and 47.14% of them use common towel for all cows, 30% of them use separate towel each cow and 22.86% of them never used towel to dry the udder during milking (Table 4).
Table 4: Hygienic condition of cows and milkers (N=70).
 Practices by milk retailers in sale and Storage of Milk: About 85.5% of the milk vendors reported as they bought milk from different farmers in their districts while restaurants were supplied (47.73%) by district farmers and (40.91%) by vendors. Most of the customers of vending shops were households and neighbouring restaurants while individual people were the major customers of restaurants. More than 81.5% of vendors sold raw milk but milk was sold after direct boiling in most restaurants. The cleanness of vendors and their surrounding environments in each districts was dirty as compared to restaurants (Tables 5 & 6).
Practices by milk retailers in sale and Storage of Milk: About 85.5% of the milk vendors reported as they bought milk from different farmers in their districts while restaurants were supplied (47.73%) by district farmers and (40.91%) by vendors. Most of the customers of vending shops were households and neighbouring restaurants while individual people were the major customers of restaurants. More than 81.5% of vendors sold raw milk but milk was sold after direct boiling in most restaurants. The cleanness of vendors and their surrounding environments in each districts was dirty as compared to restaurants (Tables 5 & 6).Table 5: Cleaning practices, source and sale of milk by milk vendors (N=32).
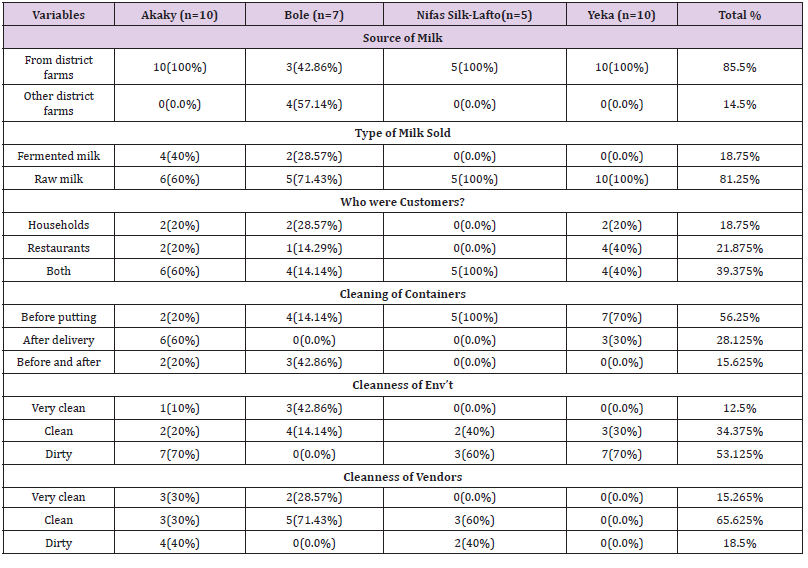 Table 6: Source, sale and preparation of milk by restaurants (N=44).
Table 6: Source, sale and preparation of milk by restaurants (N=44).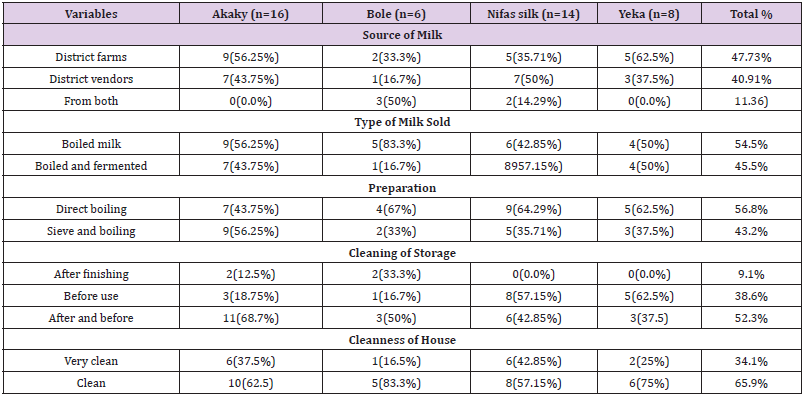 Microbiological Quality
Microbiological QualityBacterial Load Determination: From the total of 146 milk samples, 40 (29 raw and 11 boiled) milk samples were analysed for bacterial contamination by using total aerobic bacterial count (TABC) and total coliform count (TCC). The results revealed an overall mean of 7.17 log10 cfu/ml for TABC and 4.85 for TCC with more counts found in raw milk (Table 7).
Table 7: Mean comparison of total aerobic bacterial counts and total coliform counts (log10cfu/ml) among sample sources and between sample types.
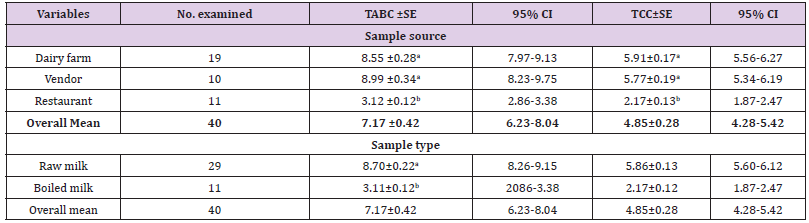 a. Total aerobic bacterial count (TABC): The results of TABC for milk from dairy farms, from vendors and from restaurants were 8.55, 8.99, and 3.12 log10cfu/ml respectively (Table 7). The mean TABC of raw milk was 8.70 log10 cfu/ml while the mean TABC of boiled milk was found 3.12 log10cfu/ml. There was no statistical significant difference between mean TABC of farms and vendors milk. However, significant difference was observed between mean TABC of dairy farm and restaurant milk, between dairy farm and restaurant milk, (Table 7) and also between raw and boiled milk (Table 7).
a. Total aerobic bacterial count (TABC): The results of TABC for milk from dairy farms, from vendors and from restaurants were 8.55, 8.99, and 3.12 log10cfu/ml respectively (Table 7). The mean TABC of raw milk was 8.70 log10 cfu/ml while the mean TABC of boiled milk was found 3.12 log10cfu/ml. There was no statistical significant difference between mean TABC of farms and vendors milk. However, significant difference was observed between mean TABC of dairy farm and restaurant milk, between dairy farm and restaurant milk, (Table 7) and also between raw and boiled milk (Table 7).b. Total Coliform Count (TCC): The mean TCC of milk from dairy farms, vendors and restaurants were 5.91, 5.77, and 2.17 log10cfu/ml respectively (Table 7). About 5.86 log10 cfu/ml mean TCC and 2.17 log10cfu/ml mean TCC were counted from raw and boiled milk respectively (Table 7). There was no statistical significant difference between mean TCC of farms and vendors milk. However, significant difference was observed between mean TCC of dairy farms and restaurant milk, between dairy farm and restaurant milk, (Table 7) and also between raw and boiled milk (Table 7).
Isolation and Identification of E.coli and E.coli O157:H7
Out of 248 (102 raw milk, 44 boiled milk, 72 farm equipment swab, and 30 vendor equipment swab) samples examined for bacteriological status of E. coli and E. coli O157:H7, about 123(49.60%) and 17(6.85%) positive isolates were found respectively (Tables 8 & 9). More isolates of E.coli were found from dairy farms and vendors. With regard to sample type majority of E.coli isolates were found in raw milk samples and farm equipment swab samples followed by vendor equipment swab samples (Table 9). However, most E.coli O157:H7 isolates were found from dairy farms and vendors. Most isolates of E.coli O157:H7 were detected in raw milk. There was no positive result from boiled milk samples taken from different restaurants of each sub cities for both E.coli and E.coli O157:H7 (Tables 8 & 9). The highest and lowest isolation rate of both E.coli and E.coli O157:H7 from Akaky Kaliti and Nifas silk-Lafto sub cities respectively. The isolation rate of both E.coli and E.coli O157:H7 was associated with sample type, source and districts, and the difference was found statistical significant (p<0.05) except with districts for E.coli O157:H7 (p>0.05) (Tables 8 & 9). Antimicrobial Susceptibility of E. coli O157:H7
Table 8: Isolation rate of E. coli from different districts, types and sources of samples that indicate the bacterial status in farms, vendors and restaurants.
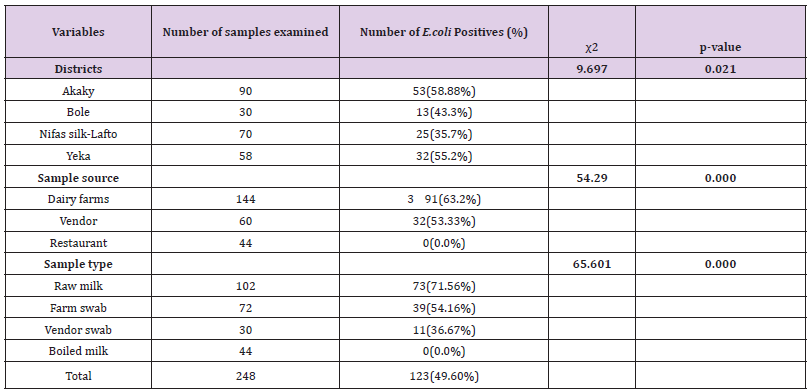 Table 9: Isolation rate of E. coli O157: H7 from the four subcities, different types and sources of samples that indicate the bacterial status in farms, vendors and restaurants.
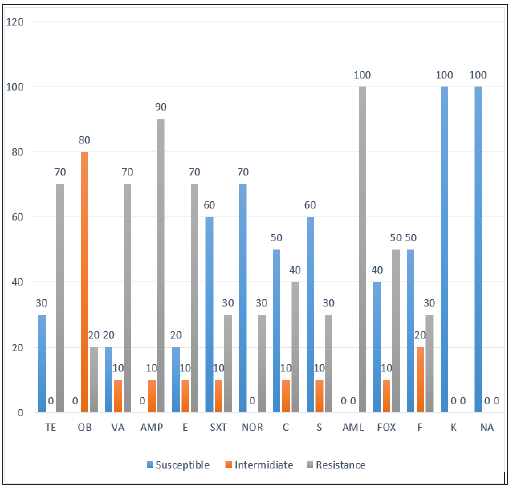
Table 9: Isolation rate of E. coli O157: H7 from the four subcities, different types and sources of samples that indicate the bacterial status in farms, vendors and restaurants.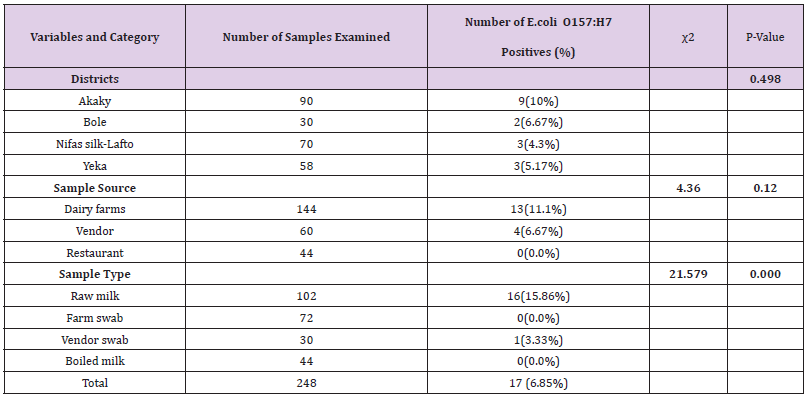 All isolates of E. coli O157: H7 were subjected to antimicrobial susceptibility test, using 14 selected antimicrobials. The isolated strains were susceptible (100%) to Nalidixic acid (NA30μg) and kanamycin (K30μg), (60%) to sulfamethoxazole trimithoprim (SXT25μg), and streptomycin (S10μg), (70%) to norfloxacin (NOR10ug), (50%) to both chloramphenicol (C30μg) and Nitrofuration (F50μg), (40%) to cefotaxime (FOX30μg), (30%) to Tetracycline (TE30μg) and (20%) to erythromycin (E15ug) and vancomycin (VA30ug) (Figure 1). From all antimicrobials used amoxicillin (Aml25μg) was (100%) resistance to all isolates followed by ampicillin (AMP10ug) (90%), tetracycline (TE30μg), erythromycin (E5ug) and vancomycin (VA30ug) (70%), cefotaxime (FOX30μg) (50%), chloramphenicol (C30μg) (40%), streptomycin (S10μg), sulfamethoxazole trimithoprim (SXT25μg), and nitrofurantion (F50μg) (30%), and cloxacillin (OB 5ug) (20%) (Figure 1).
All isolates of E. coli O157: H7 were subjected to antimicrobial susceptibility test, using 14 selected antimicrobials. The isolated strains were susceptible (100%) to Nalidixic acid (NA30μg) and kanamycin (K30μg), (60%) to sulfamethoxazole trimithoprim (SXT25μg), and streptomycin (S10μg), (70%) to norfloxacin (NOR10ug), (50%) to both chloramphenicol (C30μg) and Nitrofuration (F50μg), (40%) to cefotaxime (FOX30μg), (30%) to Tetracycline (TE30μg) and (20%) to erythromycin (E15ug) and vancomycin (VA30ug) (Figure 1). From all antimicrobials used amoxicillin (Aml25μg) was (100%) resistance to all isolates followed by ampicillin (AMP10ug) (90%), tetracycline (TE30μg), erythromycin (E5ug) and vancomycin (VA30ug) (70%), cefotaxime (FOX30μg) (50%), chloramphenicol (C30μg) (40%), streptomycin (S10μg), sulfamethoxazole trimithoprim (SXT25μg), and nitrofurantion (F50μg) (30%), and cloxacillin (OB 5ug) (20%) (Figure 1).Figure 1: Antimicrobial susceptibility pattern of E. coli O157: H7.
 Discussion
DiscussionThis study aimed at assessing the milk handling practices, possible risk factors for microbial contaminations, and clarifying the involvement of E.coli and E. coli O157:H7 as important milkborne pathogens by using laboratory analysis. This was due to the fact that milk produced in Ethiopia by many dairy sectors is not well regulated and such milk may pose a health hazard due to contamination with pathogens. Generally, it was found that, preand post-harvest handling practices of milk, prolonged storage, transportation and retailing predisposed the milk to microbial contamination. The general hygiene of milking is known to affect the numbers of microorganisms in the milk. It is recommended that before milking, the animal house should be cleaned; the udder should be washed and dried before milking. After milking, teat dipping with suitable disinfectant is necessary to control entry of microorganisms through the teat canal [18]. In the current study it was observed that milking was done in the cowsheds with dirty floor, slope and roof in most dairy farms.
This could be another risk practice that contributed to high microbial contamination of milk from farmers. [19] farmers milking in open air exposure to contaminants enter from the environment. [20] also who reported farmers milked their animals from undesignated poorly maintained milking shades or parlors predisposing milk to contamination and spoilage. Maintaining the sanitary condition of milking area is important prerequisite for clean milk production [18]. Most of the respondents 74% clean their barn daily while 26% were clean three times a week. This result is different with the result of [21] who reported about 47% of the respondents in Gurage Zone, Ezha district clean their barn three times a week. During this study, more than 77.5% of farmers did not clean their hands between milking and also milk producers and milk collectors in the study areas did not cover their hair and dressing gown during milk collection.
Indeed, the hand milking using unwashed hand practiced by famers may indicate that microorganisms on hands could result in contamination of the milk. The present finding is comparable with the result reported by [22] about (69.4%) of dairy producers in Adea Berga and Ejerie districts washed their hands between milking while the rest 30.6% did not wash their hands. Cleaning of the udder of cows before milking is one of the most important hygienic practices required to ensure clean milk production. This is important since the udder and teats of the milking cows could have direct contact with the ground, urine, dung and feed refusals and increase bacterial counts [23]. As observed in this study, 74.3% of the dairy farmers washed their cow’s udder (teats) before milking and 25.7% were not washing. [22] reported related result in Adea Berga and Ejerie districts where 62.2% of the dairy producers washed their cow’s udder before milking and 37.8% were not washing. The current result was lower than [24] reported that 82.5% of the small size farm owning households in Hawassa city practice pre milking udder washing. Contrary to this result [21] who reported that all respondents in Gurage Zone, Ezha district did not use udder washing before milking.
The use of individual towel and following essential cleaning practices during milking is important for the production of quality milk [18]. However, about 22.86% of the dairy farmers did not use towels for udder drying, 47.14% used common towel and 30 % reported as they used individual towel for udder drying. Milking in dry condition significantly reduces bacterial count. It is because no surplus water remains in the surface of the udder to drip into the milk and due to less chance of leaching dirt and bacteria from udder, teats and hands into milk [25-26] reported that thorough cleaning of the udder followed by drying with a clean cloth was effective in reducing the number of bacteria in milk contributed from soiled teats. Most of the farmers were used plastic made milk containers during milking and transported the milk to vending shops. [21] reported similar result in Ezha district of Gurage Zone where all farmers used plastic jars as milking utensil. [22] also reported similar findings in Adea Berga and Ejerie districts as all farmers used plastic milk containers during milking and transportation. Milk containers such as non-food grade plastic cans, buckets and jar cans are not recommended in the production of clean milk [23].
Aluminum containers are recommended because they don’t have adhesive properties and therefore easy to clean when compared with plastic containers [27]. Milking and milk storage utensils should properly cleaned and maintained. In addition, storage and handling of milk under room temperature increases bacteria multiplication. Therefore, cleaning and disinfections of equipment after each milking is important for reduction of milk contamination from the equipment [28]. Producers should pay particular attention for the type as well as cleanliness of milk equipment. In the present study, most of the dairy producers, milk vendors and restaurants workers washed milking utensils before and after every use. About 52.86% of dairy producers were cleaned their milking utensil before and after usage. About 76% of the respondent washed their milk container with cold water and soap while 24% used hot water and soap
This was almost similar with the finding of [22] who reported 77% of the respondent washed their milk container with cold water and soap while 23% used hot water and soap. However, the current finding contradicts with the finding of [24] who reported about 85.6% of the producers used warm water together with detergents to wash milk handling equipment while 12.1% of them cleaned with cold water. Bacteriologically, high TABC and TCC were encountered in most of the samples which were above the recommended East Africa Community standards [29]. Bacterial load in milk indicates the degree level of hygiene practiced in the whole milk production process. A total aerobic bacterial count is an indicator for prolonged storage of milk especially when stored at room temperature. The results of the present study showed an overall mean TABC of 7.17 log10cfu/ml with more counts reported in milk from vendors. The mean TABC of milk from farmers and vendors were above the required standard implying poor microbiological quality. The presence of such high bacterial load, without significant difference between vendors and farms (p>0.05), may not be surprising since the untreated raw milk harvested from dirty animals; dirty animal houses, the unhygienic environment and general milk handling might contaminated the raw milk.
The results of this study are parallel with other studies done elsewhere in Ethiopia by [22,30,31,21,33,18] in which most of the samples tested had higher bacterial count above standards. These findings are also comparable with studies done in Tanzania by [34- 37] and in Ghana by [38], but differ from the study done in Sudan [39]. The overall mean of TCC was found to be 4.85 (log10cfu/ml) with more counts recorded in farm but there was no significant difference (p>0.05) between vendors and farms. The higher coliform count found in the current study might be attributed to the initial fecal contamination of the milk through the udder, milkers, milk containers and milking environment. The coliform count obtained in the present study is similar with the result reported by [22] but higher than that reported by [30,31,21,33,18]. Since it is not practical to produce milk that is always free of coliforms, even at high level of hygienic condition; their presence in raw milk to a certain extent may be tolerated. Meanwhile according to East African Community standards for CPC of raw milk [29], good quality raw cow milk should not exceed mean TCC of 3 (log10 cfu/ml). In reference to this limit, the present study also indicates unhygienic handling of milk.
Coliforms are used as indicator microorganisms and their presence suggests a risk that other enteric pathogens may be present in the milk and implies poor hygiene. The presence of coliforms therefore indicates a safety risk, and the numbers should therefore be of the minimum recommended levels in milk products. Earlier studies by [40,41] reported that poor housing conditions can be source of contamination of coliforms for housed cows, mainly from bedding material which are mixed with cow dung and urine. Such environment contaminate teats, tail and other body surfaces from which microorganisms gain access into milk during milking. Contamination of bedding materials can be very high due to absorption of urine and faeces. Also as it was observed during this study they used untreated tap water during milking. The use of safe or boiled and portable clean water with detergent in washing milking equipment, hands and udder is a good way to remove milk leftovers including pathogens which affect the microbiological safety of raw milk [42]. Interestingly, the present study revealed that, the mean levels of contamination in boiled milk (3.12 log10 cfu/ml for TBC, and 2.17 log10 cfu/ml for TCC) which is below the acceptable standards and most likely safe for consumption. There was also significant difference between raw and boiled milk (p<0.05).
In Ethiopia, raw milk in value chain is commonly distributed locally to consumers with no controlled measures to maintain the quality and safety before it reaches consumers. This study revealed that raw milk collected from vendors and restaurants was already contaminated at the farm level. Evaluation of E. coli in raw milk is important because of the fact that the bacterium is used as an indicator organism for faecal contamination. Overall isolation rate of 123 (49.60%) of E. coli was detected from the majority of raw milk and equipment swab samples, but not detected in boiled milk. Difference in isolation rate between districts, sample sources and types was found statistically significant (p<0.05). Isolation of E. coli observed in this study was lower than 83% reported in Dares Salaam, Tanzania [43] and 100% reported in Tanga, Tanzania [44]. However, it was higher than reported in Addis Ababa, 18.6% by [45] and elsewhere in Ethiopia 15.89% by [46], 22.2% by [24] and 4.2% by [47], 11.20% in Ghana [38] and 23% in Botswana [48]. This higher isolation rate could be due to poor hygienic handling practices and initial contamination from unhygienic environment of farms and vendors.
The overall isolation rate of E.coli O157:H7 was 17(6.85%) suggesting that there is high contamination rate. Even though no isolation of E.coli O157:H7 was not recorded in boiled milk, it was found higher in raw milk which endangers the health of raw milk consumers. Most the isolates was found from dairy farms followed by vendors meaning that the contamination had originated from animals. However, it should be noted that the milk was pooled from a bulk collection hence the findings could not reflect the status of individual cow. With such a high isolation rate of E.coli O157:H7 in milk poses a threat to milk consumers. Although the isolation rate of E.coli O157:H7 showed significant difference with regard to sample sources and types (p<0.05), it didn’t reflect any significant difference between districts (p>0.05). This could be due to similar management practices, marketing chain as some of farmers sold their milk to other district vendors/shops. Higher isolation of E.coli O157:H7 was reported 8% in Ethiopia [47], 8.3% from Iran [49] and 9.6% from Iran [51]
However, [50] isolated only 1% of E. coli O157:H7 in milk samples from marketing survey in the Kenyan highlands. Previous study by [44] did not isolate E. coli O157:H7 in milk. In the study by [52] reported negative results of E. coli O157:H7 in all tested samples from cattle slaughtered at Vingunguti in Dares Salaam. Again in Ghana, [38] reported negative results in all 250 milk samples tested. Antimicrobial resistance pattern of E.coli O157:H7 isolates from animal and human sources have been reported in Ethiopia by [47]. In this study, all the isolates of E.coli O157:H7 were highly susceptible to kanamycin and nalidixic acid followed by norfloxacin, sulfamethoxazole-trimethoprim, streptomycin and, chloramphenicol. This finding is in line with the work of [53,54,47]. However, the study conducted in Saudi Arabia [55], revealed that there was resistant strain to the drugs such as tetracycline, nalidixic acid, kanamycin, sulfamethoxazole-trimethoprim, and chloramphenicol.
This variation probably attributed to the expression of resistant gene code by the pathogen which associated with emerging and re-emerging aspects of the isolates with the regards of different agro ecology [56]. On the other hand, the present study revealed that all isolates were highly resistant to ampicillin and amoxicillin. Similar findings were reported by [57,58]. This might be due to the use of inappropriate antibiotics for treatment of diseases [59] and also excessive use of antimicrobials for therapeutic and prophylactic treatment [60]. In the present study, most isolates have multiple drug resistance. This finding is analogous with other earlier findings [61-64]. This multi-drug resistance occurred might be due to administration of different antibiotics for prophylaxis or infection, discriminant use of antibiotics in the farms and another possibility is that cows are being treated with antibiotics for other conditions, thereby selecting for resistant populations of E. coli O157:H7. Such multi drug resistance may apparently be occurred which may ultimately replace the drug sensitive microorganisms from antibiotic saturated environment [65]. In conclusion, milk produced by farmers and supplied to milk vendors and restaurants in the study area contains unacceptable levels of hygiene indicators and with a potential source of milk-borne pathogens and raises a public health concern about its safety. Most of the milk supplied to the consumer in the city was managed under poor hygienic condition at ambient temperatures with poor levels of sanitation in plastic containers. The quality of water used for cleaning purposes (to wash the udder, milk equipment, and hands), however, is not secured.
Most of the producers were managing the raw milk with limited awareness and knowledge about milk contamination and public health impact of milk-borne pathogens. The hygienic conditions vary according to the production system, adapted practices, level of awareness, and availability of resources. Since the milk is managed at an ambient temperature, high microbial populations can be reached within short period of time. The sources of E. coli in the raw cow milk may be from contaminated udders, contaminated water, poor sanitation practices, contaminated containers, and milk handlers. The presence of E. coli as an indicator organisms and E. coli O157:H7 as pathogenic organisms in raw milk samples showed that the milk is of poor microbiological quality and of public health risk. E. coli O157:H7 can cause infection as well as toxic infection at a very low infective dose. The study also indicated that the E. coli O157:H7 isolates were resistant to most of the antimicrobials which may aggravate E. coli O157:H7 infections in the future. Therefore, Training and awareness creation is needed for dairy farmers, milk retailers and the public about milk handling practices and the risk of milk borne diseases.
Quantification and Modeling of Biological Processes for Tissue Engineering and Regenerative Medicine-https://biomedres01.blogspot.com/2020/11/quantification-and-modeling-of.html
More BJSTR Articles : https://biomedres01.blogspot.com



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.