A Study of Virulence Factors Associated with ESβL Producing Escherichia Coli Isolates from Patients with Urinary Tract Infection at A Nigerian Tertiary Hospital
Introduction
Urinary tract infection (UTI) is a term that describes all infections associated with the urinary system. Clinically, it is detected by the presence of significant levels of bacteria count in the urine (hence “bacteriuria”) usually 104-106 colony forming units per milliliter (cfu/ml) [1]. Different organisms have been implicated in the etiology of UTIs, the most commonly encountered being Escherichia coli, which accounts for over 80% of community acquired infections [2] and responsible for about half of nosocomial UTIs [4]. Generally, the success of any pathogen depends on its ability to
i. Enter the host,
ii. Bypass host defenses,
iii. Colonize and cause infection,
iv. And resist elimination by antibiotics as well as immunological factors created by the host and ensure effective transfer to another host.
The ability to accomplish (i)- (iii) and part of (iv) has been found to be the result of an interplay of virulence factors, most of which have been studied in detail [5]. Previous reports have shown that there are no particular virulence characteristics associated with UPEC, although it has been found to express an array of common virulence traits, hence its simple definition by Hilbert as any E. coli isolate found in the urine of a patient with UTI [3]. This phenotypic and genomic diversity observed in UPEC as an extra-intestinal pathogenic E. coli (ExPEC) [6] suggests that the pathogenic basis of success of UPEC varies from strain to strain, a hypothesis that brings in the possibility of factors other than virulence being responsible for the success of these pathogens.
Studies in recent years have reported UPEC to harbor multidrug resistance genes, with a marked association with Extended spectrum β-lactamase [ESβL] genes [7]. ESβL confers resistance to third generation cephalosporins and monobactams and are often borne on large plasmids (80kb or larger) which simultaneously harbour genes that code for multi-resistance to several groups of antibiotics [8]. ESΒLs have evolved as mutated variants of the TEM and SHV type of β-lactamases, which were the first plasmid mediated β-lactamases to be reported [9]. Today, several ESBL types have been discovered and described [10], and more are still being discovered. The hypothesis of this study is that multidrug resistance (in this case ESβL) rather than the traditional urovirulence parameters in UPEC may explain the success of these pathogens.
Methods
The isolates used in this study were isolated from the urine of UTI patients at the Obafemi Awolowo University Teaching Hospitals Complex (OAUTHC), Ile-Ife, Nigeria among other organisms. The antibiotic resistance patterns and presumptive ESβL status had been investigated and published in an earlier article [11]. Of the 41 E. coli strains isolated in the initial study, 36 were recovered and used in this study. Tests for selected virulence factors, antibiotic susceptibility and phenotypic ESβL screening were conducted according to standard references [12]. Further screening for genes that code for ESβL production were conducted using the polymerase chain reaction and the amplicons obtained subjected to standard agarosegel electrophoresis.
Antimicrobial Susceptibility Testing
Susceptibility to nine antimicrobial agents namely nitrofurantoin (300μg); ofloxacin (5μg), ciprofloxacin (5μg), gentamicin (10μg), chloramphenicol (30μg), trimethoprim (5μg), cefotaxime (30μg), ceftazidime (30μg), and sulphonamide (250μg) (Abtek, Liverpool) was tested by the disc diffusion technique according to the guidelines by the Clinical and Laboratory Standards Institute (CLSI): Briefly, four or five colonies of each test organism taken from a nutrient agar culture plate were inoculated into 10ml of sterile water using a sterile loop. The suspension was thoroughly mixed with a spin mixer. The resulting suspension was adjusted to a turbidity of 0.5 McFarland standard (A625nm =0.09). This was then applied to the surface of over-dried Mueller Hinton agar and spread evenly with a sterile cotton tipped applicator (Sterilin Ltd, Middlesex, UK). The inoculated plates were incubated at 37oC for 20 minutes for acclimatization and growth of the inoculums. Antibiotic discs (Abtek, Liverpool, UK) were then lightly but firmly pressed onto the surface of the plates equidistant to each other using a pair of sterile forceps and the plates were refrigerated at 4oC for 30min to ensure adequate diffusion of antibiotics. E. coli ATCC 25922 was used as control strain. All plates were incubated at 37oC for 18h. The diameters of inhibition zones were measured in millimeters and interpreted according to CLSI recommendations.
Extended-Spectrum Β-Lactamase Screening Tests
ESβL production was detected using the disc diffusion technique. The test plates were inoculated as for a standard disc diffusion test. Discs containing expanded spectrum cephalosporins (ceftazidime, 30μg; cefotaxime 30μg, Mast group, Merseyside, UK) and their respective combinations with clavulanic acid (5μg) were applied 30mm apart (centre to centre). E. coli ATCC 25922 was used as negative control while a positive control strain- K. pneumoniae K01 previously identified as ESβL positive [21]. After overnight incubation at 37oC, the ESβL positive strains were identified by an initial resistance pattern (≤22mm for ceftazidime and ≤27mm for cefotaxime) which is offset by the presence of clavulanic acid in the combination discs, by a standard of a ≥5mm difference in the inhibition zones.
Tests for Virulence
Sorbitol fermentation: This experiment comprised of a phenol red-peptone water base and a sugar stock solution. The inocullaused were prepared by re-streaking onto fresh plates of sterile nutrient plates and incubated at 37oC for 24h. Preparation of Phenol Red- Peptone water base: 2.8g of peptone water (Oxoid) was weighed into an empty clean and dry beaker. Phenol red (0.0075g) was weighed using the Mettlers Chemical balance (Toledo, Switzerland) and added to the peptone water, after which distilled water was added to dissolve the contents of the beaker and to make up to 150mL. The resulting solution was dispensed in 4mL volumes each into empty test tubes. A Durham’s tube was put into each of the test tubes in an inverted position and sterilized by autoclaving for 15min at 121oC.
Preparation of Sugar Stock Solution
sorbitol (5g) was weighed and dissolved in 100mL of distilled water. It was subsequently sealed and sterilized by autoclaving for 15minutes at 121oC. Sterile sugar stock solution (1ml) was aseptically added to 4mLs of each of the peptone water base to make a total of 5mlin each test tube 2h before use. A single colony was picked from the already incubated plates and was inoculated in the test tube containing 5mL of peptone water sugar medium. This was done for all the isolates and ATCC 25922 was used as a control. The tubes were incubated at 37oC for 48-72h. Absence of color change as well as absence of gas indicates a positive reaction i.e., the presence of verocytotoxin.
Haemolysin Production
All E. coli isolates were tested for haemolysin production by inoculating 18h nutrient broth cultures onto nutrient agar plates containing 4% citrated whole human blood and incubating at 37oC for 18h. The plates were then observed for α or β haemolysis indicating lysis.
Exopolysaccharide Production
In order to distinguish between encapsulated and nonencapsulated strains, the combined positive and negative staining method of Okeke and Lamikanra was employed [13]. A distinct colony was removed from 48h nutrient agar culture plate of isolates and mixed with a drop of aqeous Congo red on a microscope slide and spread to form a thin film. The slide was allowed to dry and passed over a Bunsen flame to fix before flooding with 0.2M HCl. The thin film was blotted dry and stained with ZiehlNeelson’scarbolfuschin for 10-15 min. The excess dye was poured off and the slide blotted dry without rinsing. Smears were observed at x1000 magnification under the oil immersion lens of a light microscope. The isolates appeared red against a blue background, while the presence of a capsule was observed as a clear zone around each cell.
Biofilm formation
For this experiment, liquid Artificial Urine Medium (AUM) was prepared as described by Brooks and Keevil [14] so as to mimic the conditions of the urinary system which facilitates the expression of uropathogenic traits, one of which is adherence. The tissue culture plate method was employed in the test for adherence. Exactly 10 ml each of sterile AUM was inoculated with a loopful of test organism from overnight culture on nutrient agar and incubated at 37oC for 18h. The culture was further diluted 1:100 with fresh medium. Seventy-two wells of the flat bottom tissue culture plates (Greiner, UK) were filled with 0.2 ml of diluted cultures individually. Sterile medium served as negative control. Similarly, all controls were diluted and put in the wells of the plates. After incubation at 37oC for 24 h, the plate was tapped gently, and the wells were washed with 0.2ml of phosphate buffer saline (pH 7.2) four times to remove free floating bacteria. Biofilms which remained adherent to the walls and the bottom of the wells were fixed with 2%w/v sodium acetate and stained with 0.1% crystal violet. Excess stain was washed with deionized water and plates were dried properly. The optical densities (OD) of the stained adherent biofilm were obtained with a micro plate reader at a wavelength of 570 nm. The experiment was performed in duplicates. The OD value of the sterile media used as blank was subtracted from the OD values of the test organisms so as to get actual OD values of the test strains.
Determination of presence of specific ESβL genes by Polymerase Chain Reaction
PCR amplification of possible genes encoding ESβL enzymes (TEM, CTX-M) was carried out on all E. coli isolates used in this study. The PCR primers were designated by InqabaBiotec (South Africa) according to the following nucleotide sequences;
a) blaCTX-M consensus sequence: Forward sequence-SCS ATG TGC AGY ACC AGT AA
i. Reverse sequence-CCG CRA TAT GRT TGG TGG TG [15]
b) blaTEM consensus sequence: Forward sequence- ATGAGTATTCAATTCCG
i. Reverse sequence- CTGACAGTTACCAATGCTTA [16]
A small amount of culture of each isolate was taken from nutrient aga plate using a sterile culti-loop and inoculated into an Eppendorf tube containing sterile distilled water. This was boiled for 5 minutes to lyse the bacteria and separate the DNA strands before placing on ice for at least 2 minutes. The suspension was pulse centrifuged and placed on ice again until needed. The supernactant (5 μL) was inoculated into a PCR tube containing 25 μL of mastermix (InqabaBiotec, South Africa), forward and backward primers (1 μL each) and 18 μL of PCR grade water (Inqababiotec, South Africa). The PCR mixture was placed in a thermal cycler (MJ Researcher, Watertown, MA, USA) and was subjected to a 5-min denaturation step at 94°C, followed by 30 cycles of 45 s at 94°C, 45 s at 55°C, and 60 s at 72°C, and a final elongation step of 5 min at 72°C. PCR products were separated by 100-V electrophoresis in a 2% agarose gel for 30 min, after which they were stained with ethidium bromide for 30 minutes and de-stained afterwards by placing the gel in distilled water for one hour. Gel photos were taken at a wavelength value of 250 nm. Positive control strain used was a K pneumoniae, K01 previously identified as ESβL positive [17] while the negative control was a placebo mix (without any isolate).
Results
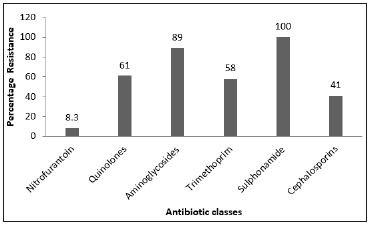
Of the 36 E. coli strains used in this study, 34 of them expressed at least one of the virulence factors tested, with bacterial encapsulation as the most significant parameter (Figure 1). All thirty-six were negative for haemolysin, while three of them tested positive for sorbitol fermentation and four were positive for biofilm formation. The antibiotic susceptibility tests results show a profile of resistance to more than one class of antibiotics. Of the 36 (100%) strains studied, 33 (91.6 %) were found to be resistant to at least three classes; 27 (75%) were resistant to at least four classes; 15 (41.6%) to at least five classes and 4 (11.1%) to six classes. As for the ESβL phenotypic screening, thirteen out of the thirty-six isolates were positive, whereas PCR test results revealed that 28 isolates carried a CTXM ESβL gene (Figure 2).
Figure 1: Chart showing the number of UPEC isolates with respect to the named virulence determinants.
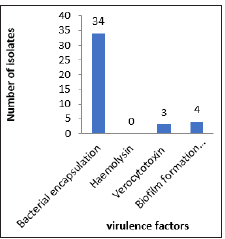
Figure 2: Chart showing the number of UPEC isolates with respect to the named virulence determinants.
Discussion
The results of this study strongly suggest that there has been trade-off between virulence and antibiotic resistance in the isolates studied, a finding that is supported by the reports of Ashley et al that UPEC which expresses little or no urovirulence traits are likely to be multi-drug resistant [18]. The issue of antibiotic resistance in the last few decades has moved from one-step mutations in the bacterial genome to the simultaneous possession of mobile genetic elements that do not only code for resistance to several antibiotics, but also function as survival tools in place of traditional virulence factors. The loss of virulence factors in favor of acquisition of multiple antibiotic resistances raises the question of continuity of the organism, in terms of its capacity to be transmitted from host to host. One possible explanation is that the ease of transmission of pathogens in the absence of traditional virulence factors has been facilitated by international trade, travel and conflicts [19]. Population explosion coupled with poor public health and suboptimal environmental hygiene practices in low- and middleincome countries account significantly for the observed high rates of transmission of infection from person to person [20,21]. The relationship between virulence and antibiotic resistance is poorly understood, the loss of virulence factors associated with quinolone resistance being the most plausible explanation for the reduction or total loss of virulence determinants observed with simultaneous multiple antibiotic resistance in bacterial pathogens [22,23].
This suggestion is further strengthened by the presence of the ESβL gene which was detected in 28 out of 36 isolates. This supports substantive research-based literature that ESβL producers are significantly more multiply resistant to antibiotics than their non-ESβL producing counterparts [24]. Furthermore, the accuracy of molecular characterization methods over and above the surface plate methods [25] of screening and investigation of microorganisms is also emphasized. The PCR amplification of blaCTXM clearly points to an ESβL phenotype, showing that CTXM ESβL is already in dissemination in this environment. The lack of information about the DNA sequences of the genes constitutes a limitation in this study, as not all blaTEM can be said to be ESβL producing unless they are sequenced [10]. A DNA sequence of the genes detected will classify the phenotypes obtained and probably explain the basis for the encountered difficulty in detecting the 15 additional isolates using the double disc diffusion method. This would give a clearer picture of the type of ESβL genes present as well as determine the presence of other β-lactamases if any, in the remaining 8 isolates in addition to the fifteen that were not phenotypically expressive.
Conclusion
Antibiotic resistance serves a multifunctional role in the survival of UPEC in this environment. The attention to the rate of occurrence of ESβL producing UPEC in this environment has also been drawn by this study, particularly when assessed from the standpoint of the extent of antibiotic use in comparison with developed countries. Current empirical antibiotic treatment choices in the country’s health care sector need to be reviewed in order to effectively manage infections caused by this group of organisms.
Who Beats the Expert? Building Precision into Simulators for Surgical Skill Assessment-https://biomedres01.blogspot.com/2021/01/who-beats-expert-building-precision.html
More BJSTR Articles : https://biomedres01.blogspot.com

No comments:
Post a Comment
Note: Only a member of this blog may post a comment.