Comparison of Pathogenic Bacteria in Water and Fecal-Source Samples in the Kathmandu Valley, Nepal, Using High-Throughput DNA Microarray
Introduction
Rapid population growth and unplanned urbanization are causing an insufficient drinking water supply, which is a major problem in the Kathmandu Valley, Nepal. People are therefore using groundwater for drinking, cooking, bathing, and laundry purposes [1]. In previous studies conducted in the Valley, various types of pathogenic bacteria, viruses, and protozoa have been detected, demonstrating the severe microbial contamination of different water sources [2-9]. Some of the possible causes of contamination for these pathogens in water bodies might be improper management of sewage, discharge of untreated industrial and hospital waste, runoff from urban surfaces, agricultural lands using animal manure, free-roaming animals, and animal farms near river banks [10,11]. A high demand for livestock products led to heavy discharge loads of feces into the environment [12]. Each gram of feces contains millions of bacterial cells, including diverse populations of bacteria [13,14]. Fecal pathogens, when discharged into the water environment, can survive for a long period, serving as a potential source and reservoir of waterborne diseases. Numerous outbreaks have been documented through waterborne transmission of fecal pathogens, resulting in mild gastrointestinal illness to even death [15-17].
In our previous study, numerous pathogenic bacteria were detected from water samples using next-generation sequencing [2]. Fecal-source samples contain many bacteria, which makes it timeconsuming to identify all pathogens and a single assay cannot be used. DNA microarray is a promising tool for the specific detection of multiple genes simultaneously [5,14]. DNA microarray has been used for molecular community analyses and microbiological surveillance studies of environmental samples, fecal sources, and human waste; thus, DNA microarray is an important tool in molecular biology [18-21]. In this study, DNA microarray targeting 941 pathogenic bacterial species/groups was used to estimate the presence of pathogenic bacteria in water and fecal-source samples. Relationships between pathogenic bacteria detected from water sources and fecal-source samples were also determined.
Materials and Methods
Collection of Water Samples and Fecal-Source Samples
A total of 16 water samples were collected from 13 shallow dug wells, a spring, a deep tube well, and a river from the Kathmandu Valley in August 2015 [22]. The sample ID for water samples were labelled as KTM. In addition, 16 composite fecal-source samples were collected from feces of two chickens, three dogs, two ducks, three pigs, three ruminants, and three sewage samples from human in December 2015 [23]. For fecal-source samples, they were labelled from initial name based on type of fecal-source except for duck. A sterile spoon was used to collect the fecal-source sample, which was then stored in a 50-mL plastic tube. Each composite sample was mixed thoroughly, and 300-400 mg of the sample was mixed with autoclaved phosphate buffered saline to obtain a 1% fecal suspension.
Bacterial DNA Extraction
Bacterial DNA from the water samples was extracted using CicaGeneus DNA Extraction Reagent (Kanto Chemical, Tokyo, Japan) as previously described [2]. A similar procedure was followed for bacterial DNA extraction from the fecal-source samples by using 5 mL of the 1% fecal suspension.
DNA Microarray Analysis
DNA microarray targeting 16S rRNA genes of 941 pathogenic bacterial species/groups, including biosafety level (BSL) 2 and 3 pathogens and other opportunistic BSL 1 pathogens listed by the Japanese Society for Bacteriology, was used. The DNA microarray used in this study was developed by Prof. Takayuki Ezaki at Gifu University, Japan, and was purchased from AMR Inc. (Gifu, Japan). The applicability of the microarray to environmental samples had been assured by the developers and previously applied to landfill leachets, river water samples, and shallow well groundwater [5,24,25]. In brief, Cy3-labeled polymerase chain reaction (PCR) products were prepared by using a PCR mixture (50 µL) containing 25 µL of SapphireAmp Fast PCR Master Mix (Takara Bio, Kusatsu, Japan), 0.25 µL each of 15 pmol/µL forward and reverse primers [5,26-28], 1 µL of DNA template, and 23.5 µL of ultrapure water. The thermal conditions were as follows: 94°C for 1 min, followed by 30 cycles of 98°C for 5 s, 55°C for 5 s, and 72°C for 10 s, and finally 72°C for 10 s. Agarose gel electrophoresis was performed to confirm the amplification of Cy3-labeled PCR products, followed by purification of the products using a NucleoSpin Gel and PCR Cleanup Kit (Macherey-Nagel, Bethlehem, USA).
DNA microarray hybridization and scanning were performed as described previously [5]. Briefly, the hybridization mixture was prepared by adding 5 µL of 10× Gene Expression Blocking Agent (Agilent, Waltham, USA) and 1 µL of 25× Fragmentation Buffer, which was mixed with 19 µL of Cy3-labeled PCR product solution. The tube was incubated at 60°C for 29 min and then cooled on ice for 1 min. Twenty-five microliters of 2× Hybridization Buffer was added to the tube and centrifuged at 15,300 × g for 1 min at 4°C. Then, the samples were hybridized with the pre-hybridized array at 55°C for 16 h. The microarray slides were washed and scanned with a GenePix 4000B (Molecular Devices, Sunnyvale, USA), and the scanned images were processed using GenePix Pro 7 software (Molecular Devices). Analysis of the scanned image data was performed as described previously [5].
Result and Discussion
Occurrence of Pathogenic Bacteria in Water Samples
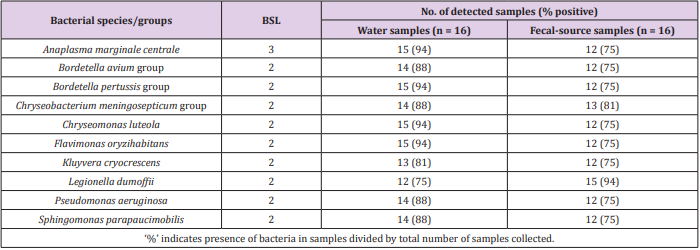
As shown in Figure 1, 31-213 pathogenic bacterial species/ groups were detected in each of the 16 water samples tested. The highest number of pathogen species/groups (213) was detected in KTM94, including 3 BSL 3 and 132 BSL 2 pathogens, followed by KTM 65 with 202 pathogen species/groups including 5 BSL 3 and 116 BSL 2 pathogens. BSL 3 pathogens were detected in 15 (94%) of the tested samples, with the highest number of 5 pathogen species/ groups in KTM65. Thirty BSL 2 pathogens and 1 BSL 3 pathogen were detected in >70% of the water samples tested. Among them, 9 BSL 2 and 1 BSL 3 pathogens were present in both water and fecal-source samples, with detection ratios of >70%, as shown in Table 1. In addition, Acinetobacter, Bartonella, Brevundimonas, Comamonas, Enterococcus, Erysipelothrix, Kingella, and Moraxella were also detected as BSL 2 pathogens, which were detected in >70% of the water samples. In the Kathmandu Valley, due to huge gap between water demand and supply, people use water sources which have not been proven to be safe. People use 12% and 22% of groundwater for drinking and cooking purpose [1]. A previous study of groundwater and river water revealed 5 BSL 3 pathogens, of which the Anaplasma marginale/centrale group (39%) was detected most frequently [27]. In this study, 6 BSL 3 pathogens were detected from different water samples, and the detection ratio of the A. marginale/centrale group (94%) was the highest (Table 1).
Table 1: Pathogenic bacterial species/groups present in >70% of both water and fecal-source samples.
A classification at the genus level revealed that 12 genera (Acinetobacter, Agrobacterium, Anaplasma, Arthrobacter, Bordetella, Caulobacter, Flavimonas, Kingella, Legionella, Moraxella, Pseudomonas, and Xanthomonas) were present in 15 (94%) of the 16 samples tested. This result was comparable to that of a previous study, where genera such as Acinetobacter, Arthrobacter, Brevibacterium, Legionella, and Pseudomonas, were detected in 82% (23/28) of water samples, which had been collected in 2014 [27]. Waterborne pathogenic bacteria that could be transmitted through the fecal-oral route were identified, whereby Vibrio cholerae, Klebsiella spp., and Salmonella enterica serovars were detected in 63%, 38%, and 31% of the water samples tested, respectively. Detection of various waterborne pathogenic bacteria in groundwater and river water samples indicates that there is a serious risk of pathogen infection through consumption of contaminated groundwater.
Occurrence of Pathogenic Bacteria in Fecal-Source Samples
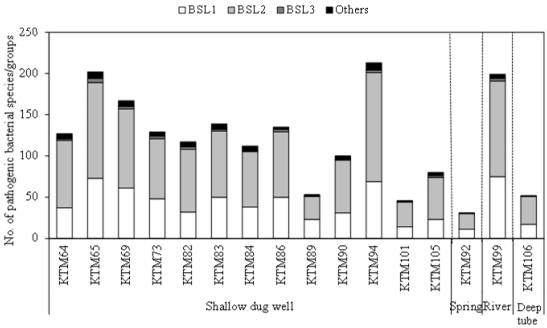
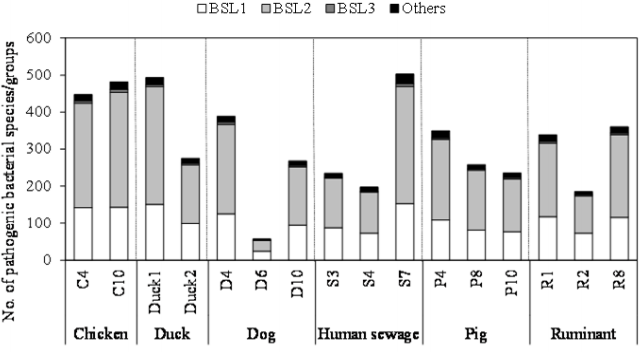
As shown in Figure 2, the number of pathogenic bacterial species/groups detected in each of the fecal-source samples ranged from 57 to 503. A total of 829 pathogenic bacterial species/groups were present in at least one of the samples tested. The highest number of pathogenic bacterial species/groups (503) was found in the human sewage S7, followed by fecal-source samples of duck (Duck1, 493) and chicken (C10, 481). Out of 611 BSL 2 pathogens targeted, 317, 316, and 310 pathogens were detected in the samples Duck1, S7, and C10, respectively. Five BSL 2 pathogens (Borrelia recurrentis, Clostridium histolyticum, C. symbiosum, Helicobacter pylori, and Suttonella indologenes) were detected in all the fecal-source samples tested. Of the total 14 BSL 3 pathogens targeted, at least one species/group was detected in all tested samples, with a maximum of 8 pathogens identified in a single sample (C10), followed by 7 pathogens in the S7 sample, and 6 pathogens in the C4 and D4 samples. Waterborne pathogenic bacteria such as H. pylori, Klebsiella spp., S. enterica serovars, V. cholerae, and Yersinia enterocolitica were detected in >60% of the fecal-source samples tested. According to the World Health Organization (WHO), at least 2 billion people worldwide use fecal contaminated drinking water, and such use is estimated to cause 500,000 deaths due to diarrhea each year [29]. In Nepal, diarrhea and gastroenteritis are the leading causes of waterborne diseases (48%), followed by typhoid/paratyphoid fever (18%) [30]. Thus, preventive management approaches are required to control the risks associated with human and animal feces.
Comparison of the Occurrence of Pathogenic Bacteria Between Water and Fecal-Source Samples
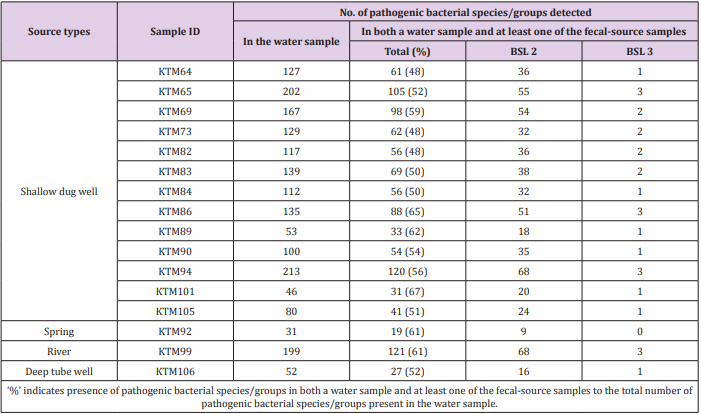
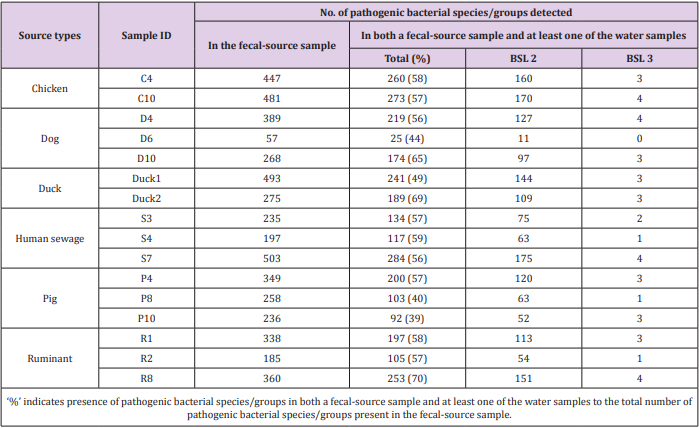
Of the 941 pathogenic bacterial species/groups tested, 361 species/groups were present in common in at least one sample each from water and fecal-source samples. The number of pathogenic bacterial species/groups that were present in both the water sample and at least one of the fecal-source samples tested ranged from 19 to 121, as summarized in Table 2. The highest detection ratio of pathogenic bacterial species/groups present in both the water and fecal-source samples was 67% (31/46) in KTM101, followed by 65% (88/135) in KTM86 and 62% (33/53) in KTM89. The highest number of BSL 2 pathogens detected in both the water and fecal-source samples was 68 in KTM94 and KTM99, whereas 3 BSL 3 pathogens were detected in KTM65, KTM86, KTM94, and KTM99. As summarized in Table 3, the number of pathogenic bacterial species/groups that were present in both the fecal-source sample and in at least one of the water samples ranged from 25 to 284. The highest detection ratio of pathogenic bacterial species/ groups was 70% (253/360) in R8, followed by 69% (189/275) in Duck2 and 65% (174/268) in D10. The highest number of BSL 2 pathogens present in both the fecal-source and water samples was 175 in S7, followed by 170 in C10, and 160 in C4. Four BSL 3 pathogens were detected in C10, D4, S7, and R8.
Table 2: Numbers of pathogenic bacterial species/groups detected in both a water sample and at least one of the fecal-source samples tested.
Table 3: Numbers of pathogenic bacterial species/groups detected in both a fecal-source sample and at least one of the water samples tested.
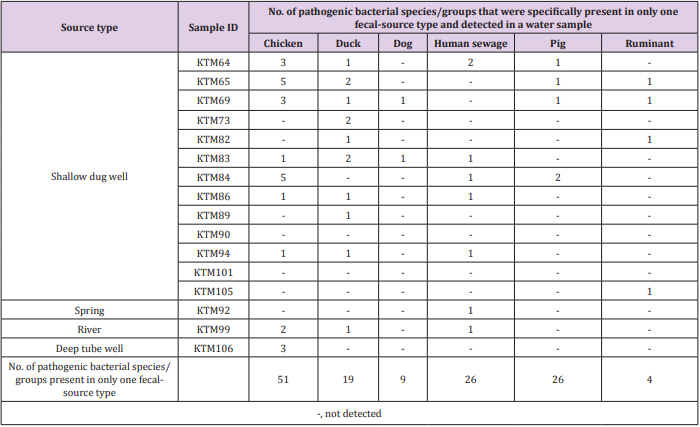
As shown in Tables 2 & 3, the detection of same species/ groups of pathogenic bacteria in both fecal-source samples and water samples implies that these bacteria in fecal-source samples might be a source of fecal contamination in water samples. Water quality, human health, and fecal-source samples are interrelated, and pathogenic bacteria in water and fecal-source samples play an important role in these three parameters [31]. Although the adverse effects of pathogens depend on the risk of exposure, environmental interaction, host susceptibility, and pathogen virulence, the effect of pathogens would be reduced if any one of these determinants are lowered [32]. Therefore, it is necessary to identify and treat species/groups of pathogenic bacteria present in environmental samples, and therefore reduce their effects on humans and animals. The numbers of bacterial species/groups detected in only one fecalsource type were as follows: 51, 19, 9, 26, 26, and 4 for chickens, ducks, dogs, human sewage, pigs, and ruminants, respectively (Table 4). The highest number of water samples with bacterial species/groups that were present in only one fecal-source sample was obtained from duck fecal-source samples (10 water samples), followed by fecal-source samples from chicken (9 water samples), and human sewage (7 water samples). In KTM69, contamination from 5 different fecal-source types was identified, whereas no fecal-source type specific contamination was found in KTM90 and KTM101. In fecal-source samples, waterborne pathogenic bacteria with detection ratios of >60%, H. pylori, Klebsiella spp., S. enterica serovars, V. cholerae, and Y. enterocolitica were also obtained in water samples with detection ratios of 6%, 38%, 31%, 63%, and 19%, respectively. Even though there are limitations in the sensitivity of probe-based assays, DNA microarray can be useful for identifying one or many sources of fecal contamination in water samples [19,33,34]. The single or multiple detection of pathogens present in both fecal-source and water samples could be efficiently performed using DNA microarray, as bacterial species/groups present only in specific fecal-source samples were also present in water samples (Table 4).
Conclusion
In summary, diverse species/groups of pathogenic bacteria, including waterborne pathogenic bacteria, were detected from water and fecal-source samples. Overall, the detection of large numbers of pathogenic bacterial species/groups by DNA microarray could be a useful tool to demonstrate interrelationships of pathogenic bacterial species/groups between water and fecal-source samples. The identification and treatment of pathogens, thus decreasing the risk of exposure to them, might reduce adverse effects on humans and animals. Therefore, safe water and public health services for prompt action should be taken to ensure that sewage is managed and treated properly before discharging into the environment, which will therefore increase the safety and quality of water for human use. Also, full sanitation is a good weapon against fecal pathogens and the sanitation can be dry sanitation and animal manure can be composted, which are not difficult in semitropical climate.
Acknowledgement
This study was supported by the Science and Technology Research Partnership for Sustainable Development (SATREPS) program of the Japan International Cooperation Agency (JICA) and the Japan Science and Technology Agency (JST), entitled “Hydromicrobiological Approach for Water Security in Kathmandu Valley, Nepal.” The authors thank Prof. Suresh Das Shrestha (Tribhuvan University, Nepal), Prof. Kei Nishida, and Dr. Takashi Nakamura (University of Yamanashi, Japan) for their continuous support in water sampling, Mr. Khadga Bahadur Shrestha (University of Yamanashi, Japan) for guidance, and Mr. Takahiro Yamada, Ms. Erina Miyata, Ms. Hitoha Moriyama, and Mr. Rui Osada (University of Yamanashi, Japan) for their invaluable support during laboratory analysis.
For more Articles: https://biomedres01.blogspot.com/


No comments:
Post a Comment
Note: Only a member of this blog may post a comment.