TP53 Promoter Methylation in EBV-Positive and EBV - Negative Gastric Cancer - A Pilot Study
Introduction
Gastric cancer is a very important problem in public health worldwide. Gastric Cancer (GC) is the fifth most commonly diagnosed cancer and the third leading cause of cancer-related mortality worldwide [1]. According to Cancer Atlas Research Network, gastric cancer is divided into 4 molecular types: GC positive for EpsteinBarr Virus (EBV), GC with microsatelite instability, genomically stable GC and GC with chromosomal instability [2]. Epigenetics has been the most rapidly expanding field in biological science in recent years. Epigenetic instability is a change in the expression of genes, not directly linked with the DNA sequence [3].
Methylation occurs primarily during the S phase of the cell cycle and is based on the covalent attachment of methyl groups (-CH3) to the nitrogenous nucleobases (around CpG dinucleotide). In the human genome, approximately 70% of CpG islands have a methyl group attached to the cytosine. However, this process does not occur in areas essential for basic processes of the cell. This includes CpG island located at the 5 ‘ends of the promoter regions of genes. Gene silencing through promoter methylation is an important risk factor in the development of different cancers. Methylation of the TP53 promoter region has been reported in several tumors. The aim of our study was to analyze the frequency of TP53 promoter methylation in EBV-positive and EBV- negative gastric cancer in Polish patients.
Material and Methods
Patients
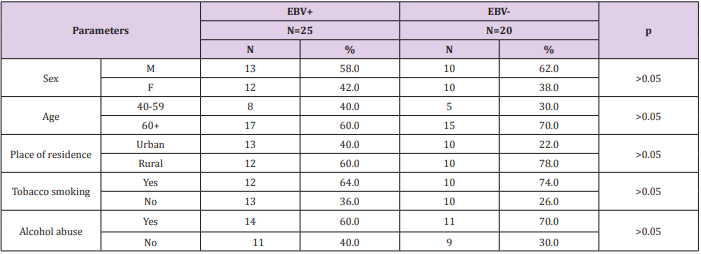
The present study consisted of a group of 45 patients with diagnosed GC, that were hospitalized at the Surgery Ward of Private Hospital in Nałęczów and AMG Hospital Center in Ryki, Poland. The patients had neither previous radiotherapy nor chemotherapy. All patients were Helicobacter pylori negative. There were no statistically significant differences between the both patient groups (age, sex, tobacco and alcohol consumption). The sociodemographic characteristics of the study group are shown in Table 1. This research was approved by the Ethics Committee and is in accordance with the GCP regulations (no. KE-0254/133/2013/ Medical University of Lublin, Poland).
DNA Extraction
The material consisted of fresh frozen tumor tissue fragments taken from patients with gastric cancer (25 EBV-positive and 20 EBV-negative). Fragments of the fresh frozen tumour tissue (20 mg) were cut and homogenized in a manual homogenizer Omni TH/Omni International/Kennesewa/Georgia/USA.
Molecular Methods
DNA extraction and detection of EBV DNA (the nested PCR) were performed as previously described [4].
Bisulfite Modification of the DNA Samples
Sodium bisulfite modification was performed using the CpGenome Turbo Bisulfite Modification Kit (Milipore, Merck KGaA, Darmstadt, Germany, katalog no S7847) according to the manufacturer’s protocol. The converted DNA was eluted and stored at – 20o C for further studies. The bisulfite-treated DNA was used as template for Methylation-Specific PCR (MSP).
Methylation-Specific PCR (MSP)
Methylation was done by MSP metod described by Herman et al. [5] and Bahtia et al. [6]. Bisulfite modified DNA was amplified with p53 gene-specific primers for methylated and unmethylated sequences: methylated: (forward) – 5’ - TTCGGTAGGCGGATTATTTG3’(reverse) – 5’ -AAATATCCCCGAAACCCAAC - 3’ and unmethylated: (forward) – 5’ - TTGGTAGGTGGATTATTTGTTT - 3’, (reverse) – 5’CCAATCCAAAAAAACATATCAC - 3’. Primer pairs were purchased from Genomed, Warsaw, Poland. Before examining the clinical samples, optimization of the reaction conditions and starters compatibility was performed using human total methylated and unmethylated DNA (CpGenome Human Methylated DNA, Milipore, Darmstadt, Germany).
Nested PCR was performed to analyze the promotor methylation status of p53 gene. PCR amplification was carried out (in duplicate for each primer pairs) in 25 µl mixture containing: water – 17.5 µl; buffer do PCR 10x - 2.5 µl; MgCl2 – 0.5 µl; dNTP – 0.5 µl; starter forward (each for reaction) 0.5 µl; starter reverse (each for reaction) 0.5 µl; polymerase Taq DNA (Qiagen) 0.125 µl; bisulfite-modified DNA 3 µl. The reaction was heated at 94°C for 3 minutes and then amplified for 45 cycles (94°C/ 45 seconds, 55°C/ 45 seconds and 72°C/ 1 minute), followed by a final 5-minute extension at 72°C. The final PCR products were analyzed on 2% agarose gel with ethidium bromide and visualized under ultraviolet light. PCR product for the unmethylated reaction - 247 bp and for methylated reaction - 193bp.
Statistical Analysis
Statistical analysis was performed to investigate the relationship between EBV presence and methylation of p53 using Pearson’s chisquare test. Statistical significance was defined as p<0.05.
Results
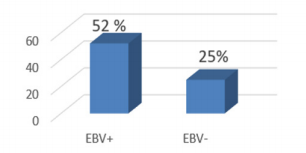
TP53 promoter methylation was detected in 13/25 (52.0%) in EBV - positive samples and in 5/20 (25.0%) in EBV – negative (Figure 1).
Discussion
Methylation of DNA is an epigenetic modification and can occur at sites where cytosine is followed by guanine. DNA methylation, particulary in promoter regions is known to silence the expression of associated genes and may occur even more frequently than inactivation of genes by mutation DNA methylation is associated with human cancers [7-9]. Most cancer-associated infectious agents, including viruses, can be divided into two categories, namely indirect and direct carcinogens. Indirect carcinogens cause cancer through chronic infection and inflammation that leads to carcinogenic mutations of the host cells [10]. Chronic inflammation is a prolonged exposure to inflammatory mediators such as free radicals, prostaglandins, and cytokines. It can cause DNA damage and oncogenic mutations in the host [11]. EBV are considered direct carcinogens because they express viral oncogenes which can directly transform host cells. During direct oncogenesis, some of or the whole viral genome integrates with the host genome and modifies the proliferation and survival pathways within the host cell [12,13].
DNA methylation patterns can be used as a biomarker for early diagnosis as well as for classification, prognosis and therapy of human cancers [12]. The promotor methylation has been found in a wide variety of genes which are involved in cell cycle, apoptosis, cell-cell adhesion, migration, invasion and metastasis [13]. TP53 gene maps on chromosome 17p13.1 and encode a tumor suppressor protein, also called p53, which takes a major role in genomic stability, cell cycle progression, cellular differentiation, DNA-repair and apoptosis. The p53 gene introduces G1 arrest in the cell cycle to allow the repair of DNA damage and prevent the cell from entering the S phase.
In our study the total frequency of the TP53 promoter methylation was identified in 18/45 (40.0%) of cases.
EBV is widespread in the world’s population (about 90%) and it is the first virus associated with a human cancer [14]. Most EBV-associated malignancies occur many years after the first infection and they are associated with latent infection. All EBV latency genes have been implicated either in cell transformation, mostly accompanied by Cp-on latency, or in oncogenesis that is primarily accompanied by Cp-off latency [15,16]. The analysis of global cellular DNA methylation detected over 13,000 differentially methylated CpG residues in cells exposed to EBV [17].
Methylation of the promotor regions is an important mechanism of silencing the genes at early stages of cancerogenesis, which is why it can be an important prognostic factor [16]. These pilot studies point to the higher frequency of methylation in EBV –associated gastric cancer than in EBV – negative tumor cases. In conclusion, the results of the current pilot study confirmed that methylation of p53 gene is frequent in gastric cancer associated with EBV infection. Increasing evidence demonstrated the use of promoter methylation of some tumor-related genes as a powerful noninvasive biomarker for the detection and diagnosis of cancer in the blood in clinical settings. We detected promoter methylation in tissue but using blood samples may be a useful biomarker in GC diagnosis and screening. For this reason, future studies in a larger group of patients are necessary. Biomarkers may provide help in the evaluation of prevention, target therapy and development of diagnostic methods of the earliest stages of tumor transformation.
Acknowledgement
This study was supported by a Research Grant from the Medical University, Lublin, Poland (DS 233).
For more Articles: https://biomedres01.blogspot.com/


No comments:
Post a Comment
Note: Only a member of this blog may post a comment.