Molecular Detection of BCL2/IGH Rearrangement in Follicular Lymphoma in Low Resource Settings: A Phase III Diagnostic Accuracy Study
Introduction
Follicular lymphoma (FL) is the second commonest non-
Hodgkin lymphoma (NHL) subtype worldwide and the commonest
in certain regions like USA [1]. FL has generally an indolent clinical
course, somehow influenced by the cytological grading that is
not, however, of prognostic relevance [2]. Conventional chemoimmunotherapy
can induce initial remissions; nonetheless, cure
is still not common [3]. In fact, relapses do occur, characterized
by progressive chemoresistance development. In a percentage of
cases, relapsing is also associated with histological transformation
to secondary DLBCL [2]. The source of relapse in patients who
initially achieve complete clinical remission are residual neoplastic
cells representing the so called minimal residual disease (MRD).
MRD can be detected either in bone marrow and blood by
molecular methods and/or in tissues (mainly lymph nodes) by PET
scan [4]. The t(14;18)(q32;q21) is molecular hallmark of FL. This
translocation joins the BCL2 gene located on chromosome 18q21
with the immunoglobulin heavy chain locus (IGH) on chromosome
14q32, leading to the inappropriate expression of BCL2 protein,
known to be a potent apoptosis inhibitor [5,6]. Detection of the
BCL2/IGH rearrangement can be clinically useful for diagnostic
purposes (using fluorescence in situ hybridization on tissues), but
also for staging and MRD monitoring (using molecular techniques
on blood and marrow) in FL patients [3,7,8].
Different techniques can be currently applied for the molecular detection of MRD, including more conventional ones (nested-PCR and quantitative Real-Time PCR, qPCR) and more innovative like digital PCR and next generation sequencing based ones [9,10]. Despite all of them have been demonstrated to be highly effective and overall reproducible and comparable [7-10], in low resource settings it is still debated whether to routinely test, due to costs, and which technique to prefer, due to technologies availability. In this study, we performed a phase 3 diagnostic accuracy study aiming to compare the two most conventional molecular techniques for MRD detection in FL, namely nested-PCR (used as test technique) and qPCR (used as golden standard) for BCL2/IGH detection. The two approaches were chosen as the only currently available in many referral centres even with limited resources.
Material and Methods
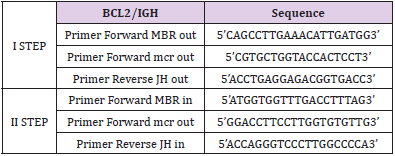
Twenty-two FL patients for which biological samples, complete clinical information, and long-term follow up were included. All patients were at diagnosis, and samples were taken before treatment initiation as well as after CHOP-R induction therapy, and after zevalin consolidation treatment at specific time-points (+3, +6, +12, +24, +30 months) [11]. Genomic DNA was extracted from mononuclear cells of peripheral blood (PB) and bone marrow aspirate (BM) as previously described [12]. The nested-PCR and the qPCR based on TaqMan technology [ABI PRISM 7900HT Fast Real-Time PCR System (Applied Biosystem)] were performed as previously reported [3,13,14]. As for BCL2/IGH PCR assays, primers were used according to previous Italian experiences [Ladetto 2001] (Tables 1-2). GAPDH was used as control gene for qPCR. Conversely, AF4 was chosen as control gene and was amplified according to BIOMED2 protocols for nested PCR [15]. All samples were tested by both techniques in triplicate.
Calculations of sensitivity (ST), specificity (SP), positive predictive value (PPV), negative predictive value (NPV), were made by CATmaker software (Centre for Evidence Based Medicine, Oxford University, http://www.cebm.net). The limit of significance for all analyses was defined as P<0.05. The study was approved by the local Ethical Committee and was developed and conducted in respect of the Helsinki Declaration. The study was designed and conducted according to the evidence-based medicine rules, respecting the STARD requirements.
Results and Discussion
All the enrolled patients could be studied for MRD. In total, 145
tests were performed. In fact, other than the expected 132 (22 cases
by 6 timepoints), additional 13 were available from patients with
longer clinical CR duration. Overall, we observed good concordance
between “qualitative” nested-PCR and quantitative real-time PCR
(80,86 %), in detecting MRD. The absolute sensitivity of the qPCR
was in line with previously reported data [7]. Particularly, by
evaluating serial dilutions of t(14;18)-positive cells into t(14;18)-
negative cells, the relative sensitivity of our qPCR assay of about 10−5
resulted greater than the nested-PCR one (10-4), with an enhanced
quantitative potential. This is overall in line with most studies. In
terms of reproducibility, the precision of qPCR was determined
by repeatability intra-assay and inter-assay; both the tests gave
results of high reproducibility, above 95% considering 3 replicates.
In contrast, the nested-PCR has given a lower reproducibility with
discordant data and the need of additional repetitions to achieve a
uniform result (three nested-PCR in mean). Overall, this is in line
with previous works on qPCR. By contrast, nested PCR seemed to
be “technically” more complicated and probably requiring more
experienced personnel, to be consistently performed. This fact,
further stress the need for adequate training and standardization
processes when MRD is studied, in order to ensure the requested
clinical consistency.
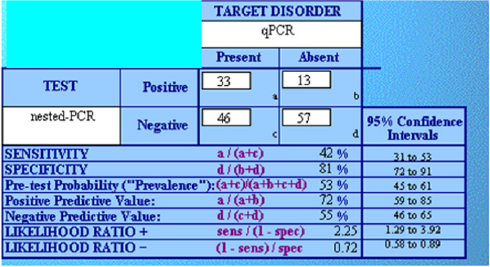
Consistency between the two was evaluated in terms of
sensitivity and specificity. Overall, this analysis confirmed what
observed in terms of reproducibility, i.e. a significantly higher
efficacy of qPCR. Among 145 performed tests, 85 were concordant
between the two techniques, while 59 were not (59% overall
accuracy). Particularly, among 103 tests turned out to be negative
by nested PCR, only 46 were instead positive by qPCR (45%).
Conversely, among the 46 that resulted positive at nested PCR, only
13 were discordant and 33 consistent (72%). This was translated
into remarkable specificity but low sensitivity of nested PCR
(Figure 1).
Lastly, analysis of costs and practical feasibility in reduced laboratories was performed. The expenses for reagents, consumables and labor employed for the TaqMan assay was calculated about 34,00€ (4,443 KES) per sample when testing the maximum number of 5 samples in triplicate in 96 well-plates. Conversely, the analysis of 5 sample by nested-PCR has a total amount of 126,00€ (16,466 KES). This calculation was obviously optimized for running a complete TaqMan plate. By reducing the number of available samples, the cost would progressively increase. This implies that referral labs centralizing the activities are advised, particularly when resources are limited, also considering the highest initial investment for machinery. The shortest test duration of 3 hours and 14 minutes was found for the real time PCR while 18 hours and 30 minutes were needed to perform a complete nested- PCR analysis (including gene control PCR, the nested-PCR repeated for three times in mean, post PCR manipulation)
Conclusion
The present study, though based on a limited series, highlights
the relevance of using a qPCR-based method to detect BCL2/
IGH rearrangements in FL patients in laboratories with limited
resources. The use of TaqMan detection system was shown to be
a sensitive, reproducible, and economical tool for MRD monitoring
in FL. It allowed a relative sensitivity of about 10-5 providing a
more accurate prognostic information [16]. Finally, the Taq Man
approach in comparison with nested-PCR showed the simplest
and shortest workflow sequence with a considerable gain of time
and money, the average cost of 34€ per samples makes it feasible
also in low resource Countries. Adequate programs of training and
standardization should be then planned accordingly.
For more Articles on : https://biomedres01.blogspot.com/




No comments:
Post a Comment
Note: Only a member of this blog may post a comment.