Integrated DNA Extraction Protocol to Avoid PCRInhibitors
from Fecal and Environmental Samples for
Next-Generation Sequencing
Introduction
The use of next-generation sequencing (NGS) techniques is ondemand
for many biological applications [1, 2]. Obtaining DNA with
good quality is crucial for any molecular technique [3] (pp 27), like
metagenomics studies [4]. High-quality DNA is referred to as DNA
with a high molecular weight, with an A260/A280 ratio between 1.8-
2.0, and without contaminant substances [5,2]. These substances
can act as polymerization inhibitors [6], the same happens with
the presence of biliary salts, urea, phenols, polysaccharides,
humic acids, tannic acid, and others [7]. These particular DNA
characteristics are hard to obtain when complex samples are
analyzed [8]. In the Biomedical Innovation laboratory at the CICESE
in México, environmental metagenomics studies are performed
regularly. Most of the time, the samples that are analyzed are unique,
unrepeatable, expensive to resample, or not typical. For example,
sea lion pups fecal cotton swabs (SPFS) from seven rookeries in the
Midriff region of the Gulf of California, were collected to perform
metagenomics of the anal-associated microbial community.
On the other hand, at the same time, several marine sediment (MS) samples from the Gulf of Mexico were analyzed in the same way (up to 3,000 m deep). These kinds of samples are unique and unrepeatable because the environmental, ecological, and physiological characteristics are in continuous change. Therefore, the metagenomics associated with these samples was unique at the time, hence the value of these samples. Even when there are some manuscripts about marine mammals metagenomics in fecal swabs [9], or about the bacterial communities present in the marine sediments; problems with the samples still exist. So, it was needed to find a way to standardize the technique for samples with these specific characteristics. For both kinds of samples mentioned, it was needed to perform a routine PCR step, which was repressed by some inhibitor. Consequently, the downstream steps for the metagenomics analyses were inhibited too. In this work, we combined the use of two commercial kits from different brands, which are relatively inexpensive and diverse use, with modifications to avoid the PCR’s inhibitors present in seal ions fecal samples, and marine sediment samples to complete the subsequent metagenomics studies.
Experimental Design
Both kinds of samples, the cotton swabs, and the marine sediment were transported in liquid nitrogen to The Biomedical Innovation Research Laboratory at CICESE, to be processed. Several DNA extractions were performed with commercially available kits to obtain functional DNA. The isolation of genomic DNA from Gram-negative bacteria SPFS was processed with two different kits: Wizard Genomic DNA Purification Kit, Promega Corporation, WI, USA, and the PureLink Genomic DNA kit, Life Technologies, CA, USA. Both protocols were modified to provide higher quality genomic DNA (section I). In the first attempt, Wizard Genomic DNA Purification kit (Promega Corporation, Madison, WI) was employed, the DNA concentration obtained was quantified in NanoDrop spectrophotometer, showing the presence of that biomolecule (Table 1), however PCR-products were not obtained. This extraction protocol was applied few more times, with modifications (data do not show), but the same results were obtained. Then, a second DNA extraction protocol was implemented, using others SPFS (PureLink Genomic DNA kit; Life Technologies, Carlsbad, CA, USA; Cat. no.: K1820-01).
Even, when the DNA concentration and A260/A280 ratio was theoretically good enough (Table 1), no PCR-products was observed in the agarose gel electrophoresis, as in the case of promega kit, different variations of this protocol were tried, but no PCR-products were obtained. For marine sediment, in the first attempt, the PowerMax Soil DNA Isolation Kit (MoBio Laboratories, Inc., Carlsbad, CA, USA; Cat. No.: 12888-50) was employed; as in the SPFS samples, despite the concentration and quality of DNA it was theoretically good enough (Table 1), no PCR-products were observed. Then, the PureLink Genomic DNA kit (Life Technologies, Carlsbad, CA, USA; Cat. no.: K1820-01), was implemented, but as in the SPFS samples, no PCR-products was observed, and as in the SPFS case, different variations were tried, for all attempts, no PCRproducts were obtained. That absence of PCR-products in both kind of samples, was related to an unknown PCR-inhibitor. To avoid the PCR-inhibitor, the two protocols were combined in a sequential way, first, Wizard protocol, then PureLink Genomic DNA kit, from the column purification step and forward. This combined protocol was named as “integrated protocol”, and then PCR-products was observed.
Once the DNA samples were standardized, the metagenomic mass sequencing analyzes with the IonTorrent NGS S5 platform (Thermo Fisher) were performed. Materials
• 10, 200, and 1000 μl wide-bore micropipette filter tips (Barrier tips, Thomas Scientific, Swedesboro, NJ, Cat. No. 1149J65, 1138W65, 1145N12).
• 100% Ethanol proof molecular biology grade (Sigma-Aldrich, St. Louis, MO, USA; Cat. No.: E7023-500ML).
• 100% Isopropanol proof molecular biology grade (Sigma- Aldrich, St. Louis, MO, USA; Cat. No.: I9516-500ML).
• Nuclease free water (Thermofisher Scientific, Waltham, MA, USA; Cat. No.: AM9930).
Materials
• Wizard Genomics DNA Purification Kit (Promega Corporation, Madison, WI, USA; Cat. No.: A1120)
• PureLink Genomic DNA Kit (Life Technologies, Carlsbad, CA, USA; Cat. no.: K1820-01).
• Ion Universal Library Quantitation Kit (Thermofisher Scientific, Waltham, MA, USA; Cat. No.: A26217).
• PowerSoil DNA Isolation kit (MoBio Laboratories, Inc., Carlsbad, CA, USA; Cat. No.: 12888-50).
• High Sensitive DNA Kit (Agilent Technologies Santa Clara, CA, USA; Cat. No.: 5067-4626).
• Ion 16S Metagenomics Kit (Life Technologies, Carlsbad, CA, USA, Waltham, MA, USA; Cat. No.: A26216).
• Ion 510 & Ion 520 & Ion 530 Kit-Chef (Life technologies, Carlsbad, CA, USA, Waltham, MA, USA; Cat. No.: A34461).
• Ion 530 Chip Kit (Life Technologies, Carlsbad, CA, USA; Cat. No.: A27764).
Equipment
• Thermoblock (ThermoStat plus, Eppendorf, Hauppauge, NY, USA; Ref. 5383000019).
• Ultracentrifuge (5418R, Eppendorf, Hauppauge, NY, USA; Cat. No.: 5401000137).
• Vortex (Benchmarck Scientific, Edison, NJ, USA; Cat. No.: BV1000).
• NanoDrop Spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA; Cat. No.: ND-LIT-PR).
• 2100 Agilent Bioanalyzer (Agilent Technologies Santa Clara, CA, USA; Cat. No.: G2939BA).
• ProFlex PCR System (Thermofisher Scientific, Waltham, MA, USA; Cat. No.: 4484073).
• Ion Chef Instrument (Thermofisher Scientific, Waltham, MA, USA; Cat No.: 4484177).
• Ion GeneStudio S5 Series for NGS (Thermofisher Scientific, Waltham, MA, USA; Cat. No.: A38194).
Samples
Sealion pup fecal swabs (SPFS): 125 cotton swabs were collected, containing the anal-associated microbial community of sea lion pups (SPFS) from seven rookeries in the Gulf of California in July-August from 2018 and 2019. Under SAGARPA permission No. SGPA/DGVS/003083/18.
Marine Sediment (MS): Recovered more than 70 deep-sea sediment samples, between 1000 m and 3700 m of depth from the Gulf of Mexico basin among 5 years of study, from 2012 through 2018.
All samples were kept in liquid nitrogen during the campaign and kept at -80ºC in the laboratory until processed.
Procedure
DNA extraction from SPFS and DFS: Wizard (Promega) protocol for Gram-negative Bacteria (Around 2.5 hours of hands-on work).
Add 600 μl Nuclei lysis Solution. Vortex 10 seconds (secs).
Incubate for 25 minutes (min) at 80°C, then cool down to room temperature. CRITICAL STEP
Add 3 μl of RNase solution. Mix and incubate at 37°C for 30 min, then cool to room temperature.
Add 600 μl of Protein Precipitation solution. Vortex 5 secs. Incubate on ice for 5 min. Centrifuge at 16000 g for 3 min.
Transfer the supernatant to a clean tube with 600μl of isopropanol. Mix. Centrifuge at 16000 g for 2 min and decant the supernatant. Carefully pour off and drain the tube.
Add 600 μl of room temperature 70% ethanol and invert the tube several times to wash the DNA pellet. Centrifuge at 16000 g for 2 min. carefully pour off and drain the tube on clean absorbent paper upside-down. Allow the pellet to air dry for 10-15 min.
CRITICAL STEP Rehydrate the DNA pellet in 50 μl of nucleasefree water and let it sit overnight at -20°C. Continue at step 3.3.
DNA extraction from MS environmental samples: PowerMax Soil DNA Isolation Kit (MoBio) (Around 3 hours of hands-on work).
Add 15 ml of PowerBead Solution to a PowerBead Tube with between 8 and 10 g of the soil sample, add 1.2 ml of Solution C1.
Vortex for 15 minutes at the highest speed.
Centrifuge tubes at 2500 x g for 3 minutes at room temperature and transfer supernatant to a clean tube, add 5 ml of Solution C2 and mix.
Incubate at 4°C for 10 minutes.
Centrifuge tubes at 2500 x g for 4 minutes at room temperature. Transfer supernatant to a clean tube, add 4 ml of Solution C3, and mix.
Incubate at 4°C for 10 minutes.
Centrifuge tubes at 2500 x g for 4 minutes at room temperature. Transfer supernatant to a clean tube, add 30 ml of Solution C4, and mix.
Fill Spin Filter with solution from the previous step and centrifuge at 2500 x g for 2 minutes at room temperature.
Discard flow-through and add a second volume of supernatant to the same Spin Filter and centrifuge at 2500 x g for 2 minutes at room temperature. Discard flow through and repeat until the entire volume has passed through the spin filter.
Add 10 ml of Solution C5 to Spin Filter and centrifuge at 2500 x g for 3 minutes at room temperature. Discard flow-through.
Centrifuge Spin Filter at 2500 x g for 5 minutes at room temperature. Place Spin Filter in a new Collection Tube.
Add 5 ml nuclease-free water to the Spin Filter and centrifuge at 2500 x g for 3 minutes at room temperature.
Store the DNA at -20°C. Continue at step 3.3.
Second phase extraction (Purification). Using the PureLink Genomic kit (Life Technologies) protocol for gram-negative bacteria (around 1 hour of hands-on work).
Add 40 μL RNase A and vortex. Add 400 μL PureLink Genomic Lysis/Binding Buffer and vortex it again.
Add the lysate sample (≈700 μL) to the PureLink Spin Column. Centrifuge at 10000 g for 1 min at room temperature. Discard the collection tube and place the spin column into a clean collection tube.
Add 700 μL Wash Buffer 1 with ethanol to the column. Centrifuge at 10000 g for 1 min. Discard the collection tube and place the spin column into a clean collection tube.
Add 700 μL Wash Buffer 2 with ethanol to the column. Centrifuge at 10000 g for 1 min. Discard the collection tube and place the spin column alone into a clean tube. Centrifuge at 16000 g for 30 secs. Place the column into a sterile 1.5 μL tube.
Add 50 μL of nuclease-free water to the column. Incubate at room temperature for ≈2 min. Centrifuge the column at 16000 g for 1.5 min.
Store the DNA at -20°C until used.
PCR conditions to test functional DNA.
This PCR was performed using the GOTaq ® DNA Polymerase reagents (Promega), with this protocol.
Add 10 ng DNA extracted
In a sterile, nuclease free microcentrifuge tibe, combine the following on ice:
4 μl 5X Green Buffer
2.4 μl 1.5 mM MgCl2
0.4 μl 10 mM dNTP´s mix
0.4 μl 0.25 μM forward primer
0.4 μl 0.25 μM reverse primer
0.1 μl GoTaq® DNA Polymerase (5u/μl)
Mix on vortex by 5s
The PCR programme consisted of an initial denaturation at 95°C for 2 min, 30 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, and a final incubation at 72°C for 5 min.
Products were resoved by agarose 1.5% (Sigma-Aldrich, A9539G)
Results
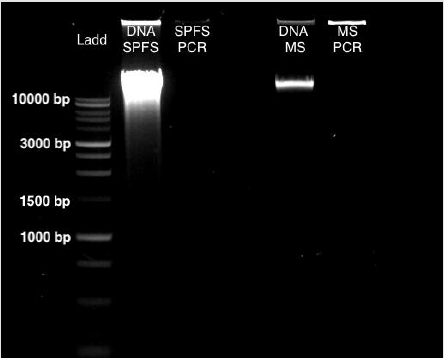
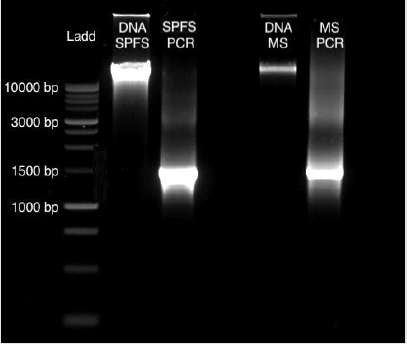
From sea lion samples, only three pooled SPFS samples were employed to standardize both protocols independently. In both cases, the PCR reaction came out negative (Figure 1). The same outcome occurs with the Marine Sediment samples extracted with the MoBio protocol, with no positive PCR results (Figure 1). Bacterial 16S gene amplification from the DNA obtained from SPFS, was positive using the integral protocol (Promega-Life Technologies) described above. The DNA of cotton swabs had an acceptable quality DNA ratio (A260/A280= 1.95) (Table 1) [5]. It was comparing the DNA concentration obtained with the separated protocols at the first stage, which presented a higher concentration with lower quality DNA (Table 1). After the integrated protocol was implemented, a final DNA concentration of 35 ng/μl for the SPFS (Table 1) and 77 ng/μl for the Marine sediments (Table 2) was obtained, with much better quality for both sample types. DNA integrity and 16S PCR products were resolved by gel electrophoresis (TRIS-agarose 2%) for the SPFS and the MS (Figure 2).
Figure 1: Gel Electrophoresis (TRIS-agarose 2%) for DNA the extractions and PCR amplification, using the Wizard Protocol (Promega) for the SPFS and the PowerMax Soil DNA extraction kit (MoBio) for the MS samples.
Figure 2: Gel Electrophoresis (TRIS-agarose 2%): DNA obtained from the cotton swabs and the marine sediments obtained with the integral protocol, with their positive PCR products of 1469 bp.
Table 1: DNA concentration and A260/A280 ratio for the Wizard Genomic DNA Purification Kit and the PureLink Genomic DNA kit protocols separately and using the integrated protocol implemented on this work.
Table 2: Marine sediment DNA concentration and A260/A280 ratio, for the Power Soil DNA Isolation kit protocol and the integrated protocol.
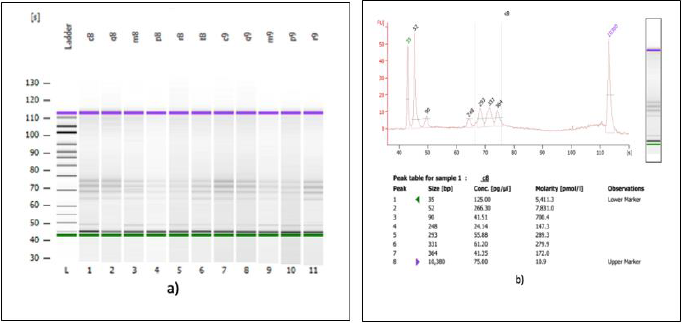
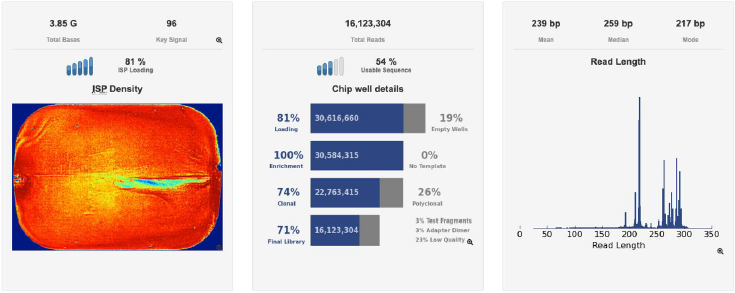
A second verification of the quality of both DNA extractions was performed using a Bioanalyzer (Agilent Technologies) (Figures 3a & 3b) and corroborated with a successful metagenomic sequencing with the IonTorrent S5 next-generation sequencing platform using the 16S Ion 530 Chip Kit (Life Technologies, Carlsbad, CA, USA (Figure 4). It is not the main of this paper to show the results of the mass sequencing mentioned, but the evidence of the successful sequencing can be seen in Figure 4. Discussion The integrated DNA extraction protocol proposed in this work is an efficient and accessible alternative for polymerization-inhibitors removal for valuable samples, or when one does not have the means or the time to identify the specific inhibitors present in the sample. It was not the subject of this work to elucidate the nature of the inhibitor or inhibitors present in the samples, but to remove any contaminant that could damage or outperform the sample analyzes.
Figure 3: Analysis of 16S genomic DNA amplification with a bioanalyzer spectrophotometer.
(a) Gel imaging of the genomic DNA extracted with the integrated protocol from 11 SPFS sample pools.
(b) Trace of one of the samples shown in figure 3a. The samples analyzed here were used for library construction and sequencing on the IonTorrent S5 platform.
Figure 4: Sequencing information of one of the chips used to sequence the samples standardized on this study, showing the total reads, chip loading, and fragment length in base-pairs, in an Ion Torrent S5 genetic sequencer (Thermofisher Scientific).
Due to the different nature of all the samples analyzed in this study, it is possible to infer which group of inhibitors were responsible for the PCR blockage. In the case of the sea lion pup’s fecal swabs, the presence of complex polysaccharides has been reported [10,11]. For the marine sediment samples, the presence of humic acid is familiar [12], which is known to inhibit the PCR reaction [7]. Without precise knowledge about the chemistry of the reagents used by the Promega and Life Technologies kits, it is only possible to theorize why the coupling of both protocols manage to eliminate or minimize the concentration of the possible inhibitors in this kind of biological samples. When the protocols are used individually, the relatively large quantity of extracted DNA obtained with the Promega protocol might have an equal proportion of the unknown inhibitor, blocking of the PCR reaction. In respect to the Life Technologies protocol, possibly the nature or concentration of the inhibitor saturates the column matrix, and it allows this inhibitor to pass through. Nevertheless, when we use the Promega protocol first, their chemistry might pre-wash the DNA, diminishing the concentration of the inhibitor, allowing the Invitrogen reagents and extraction column to purify the DNA.
This protocol employs reagents of two commercial kits that are common in most laboratories, and they are not too expensive. The results obtained here show that although there is a decrease in the DNA extraction quantity, this final DNA meets the requirements necessary to perform NGS analyzes successfully, making this coupled protocol a perfect alternative to be used in complex biological or environmental samples.
For more Articles on : https://biomedres01.blogspot.com/





No comments:
Post a Comment
Note: Only a member of this blog may post a comment.