Serum Calcium in Relation to COVID-19 in Asian Population: A Two-Sample Mendelian Randomization Study
Introduction
On February 11, 2020, the Director-General of the World Health Organization Tan Desai announced in Geneva, Switzerland, that the 2019 pneumonia infected by the new coronavirus SARSCoV- 2 was named “COVID-19” [1]. Since December 2019, the new coronavirus is still spreading widely around the world. As of 0:08 on October 13, 2021, Beijing time, there have been a total of 237,655,302 confirmed cases of new coronary pneumonia worldwide, and a total of 4,846,981 deaths, which has brought huge disasters to society [2]. At present, there is no effective medical method to kill this virus. Therefore, it is very important to prevent the new coronavirus infection. In recent years, with the vigorous development of nutrition, the role of trace elements has gradually been widely valued by the scientific community. Trace elements are active substances involved in the intermediate links of the body’s metabolism, such as calcium. Calcium is the most abundant inorganic element in the human body [3].
It has the following biological functions [3]:
1. Osteogenesis and blood coagulation;
2. Regulating cell function;
3. Acting as a messenger;
4. Helping to regulate enzyme activity,
5. Maintaining neuromuscular excitability,
6. Reduce the permeability of capillaries and cell membranes, prevent exudation, and inhibit inflammation and edema. At present, many studies have proved that serum calcium concentration is potentially related to the onset and prognosis of new coronary pneumonia. Some studies have shown that a lack of serum calcium concentration increases the risk of new coronary pneumonia infection; but some opinions are that there is no correlation between serum calcium and the incidence of new coronary pneumonia. The conclusions are inconsistent. And there is currently no research on the masses of Asians. In traditional observational epidemiology, the association between exposure and outcome may be affected by confounding factors and reverse causal associations, which limits its use in causal inference. Mendelian randomization (MR) uses single nucleotide polymorphisms (SNPs) as instrumental variables (IVs) to infer the causal association between exposure and outcome, which can overcome confounding factors and the influence of reverse causality on causal inference [4,5]. Therefore, this study uses the 2SMR method to conduct a secondary analysis of genome-wide association study (GWAS) data to explore the causal relationship between serum calcium concentration and the risk of COVID-19.
Methods
Study Subjects
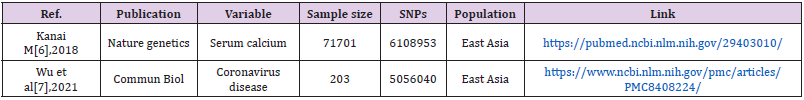
Material Source: Serum calcium concentration is an exposure factor, and whether one person is infected with new coronary pneumonia is the outcome. Download the serum calcium and GWAS summary data of whether or not the new coronary pneumonia is infected through the MR base database and the GWAS Catalog database (Table 1).
Screening of Instrumental Variables: The basic requirements for genetic variation to become IV are summarized below. The IVs is related to exposure; variation has nothing to do with confusion issues related to exposure and outcome; and variation has no effect on the results, unless it is possible to expose through contact. First, using the whole genome information of Asian populations as a reference, SNPs with a significant level (P < 5*10-8) associated with adults up to the whole genome were screened out. In genetics, it is generally believed that genetic sites that are very close on chromosomes are “bundled” together and passed on to offspring. This also results in a large r2 between sites that are very close. In order to ensure the independence between SNPs, this study set the linkage disequilibrium parameter (r2) to 0.001 and the genetic distance to 10000kb. That is, remove all SNPs whose genetic distance from the top SNP is r2 > 0.001 within 10000kb. Secondly, in the outcome data set, if the information about the filtered instrumental variable is not found, the SNiPA website is used to search for alternative SNPs (r2 > 0.9); if alternative SNPs cannot be obtained, the instrumental variable is eliminated. After that, align the allele effect values in the exposure data set and the outcome data set, and merge the data [6]. Finally,
1. SNPs that are statistically significant (P < 0.05) with the risk of COVID-19;
2. SNPs whose BMI with confounding factors reaches a genomewide significance level (P < 5*10-8); 3. F statistics < 10 SNPs.
Statistical Analysis
1. Weak Instrumental Variable Test: When the degree of correlation between instrumental variables and exposure factors cannot reach the significance level of the whole genome, or when instrumental variables can only explain a small part of phenotypic variation, such instrumental variables are called “weak instrumental variables”. In a twosample MR study, the existence of weak instrumental variables will underestimate the strength of association and reduce the power of statistical testing [7]. Therefore, this study uses the formula 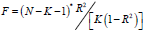 to evaluate whether there are weak instrumental variables. Among them, N is the sample size of the exposed data set, K is the number of SNPs, and R2 is the degree of variation that can be explained by the SNPs in the exposed data set [8]. The calculation formula of R2 is:
to evaluate whether there are weak instrumental variables. Among them, N is the sample size of the exposed data set, K is the number of SNPs, and R2 is the degree of variation that can be explained by the SNPs in the exposed data set [8]. The calculation formula of R2 is: 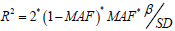 . Among them, MAF is the minor allele frequency, which is equivalent to the effect allele frequency EAF when calculating R2, β is the allele effect value, and SD is the standard deviation.
. Among them, MAF is the minor allele frequency, which is equivalent to the effect allele frequency EAF when calculating R2, β is the allele effect value, and SD is the standard deviation.
2. Gene Pleiotropic Test: When genetic variation can affect the occurrence of the outcome through other means besides exposure factors, the instrumental variable is pleiotropic [9]. The existence of gene pleiotropy will cause the “collapse” of the two-sample Mendelian randomization model. Therefore, this study used the software package “MR_PLEIOTROPY” in the R package to test the genetic pleiotropy of instrumental variables. In addition, the MR-Egger model is used to correct the bias caused by gene pleiotropy.
3. Linkage Disequilibrium Test: In genetics, it is generally believed that genetic sites that are very close on chromosomes are “bundled” together and passed on to offspring, which leads to r2 between sites that are very close. Will be very large; this phenomenon is called linkage disequilibrium. Therefore, in this study, the linkage disequilibrium parameter (r2) was set to 0.001 and the genetic distance to 10000kb to avoid the occurrence of linkage disequilibrium. That is, remove all SNPs whose genetic distance from the top SNP is r2>0.001 within 10000kb.
4. Heterogeneity Test: Cochran’s Q test is used to judge the heterogeneity of instrumental variables [10].
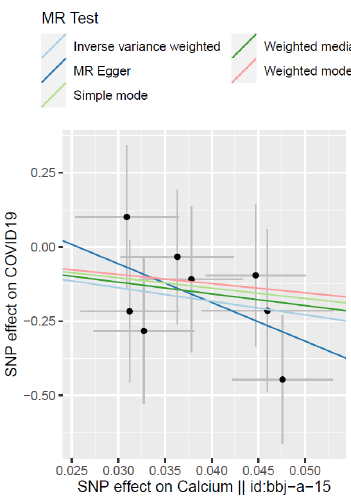
5. Causal Effect Estimation: Five models of MR Egger, Weighted median, Inverse variance weighted, Simple mode and Weighted mode are used to test the causal association between adult height and the risk of COVID-19. The result is expressed as the odds ratio (OR) of the risk of COVID-19 for each increase in height by one standard deviation. In order to strengthen the reliability of the results, the “Leave-one-out” method is used for sensitivity analysis, each SNP is eliminated in turn, and the remaining SNPs are used as IVs for two-sample MR analysis to determine that a certain SNP will have a strong causal relationship. Influence.
6. The above analysis methods were all realized by R v4.0.5 software, and the two-sided P < 0.05 indicated that the difference was statistically significant.
Results
Information About Instrumental Variables
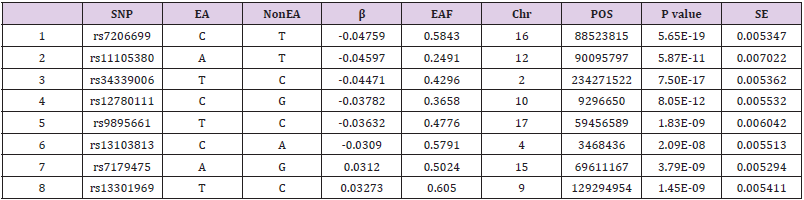
This study finally included a total of 8 SNPs as instrumental variables (Table 2). R2 is 0.25%, the F statistic corresponding to a single SNP has a distribution range of 15.32-38.50, and the F statistic corresponding to 8 SNPs is 22.32, indicating that the causal association is less likely to be affected by weak instrumental variable bias1 [11]. The MR-Egger regression intercept term is 0.33, which shows that there is no genetic pleiotropy between SNPs and the risk of COVID-19. The results of MR_PLEIOTROPY test showed that there was no gene pleiotropic bias (P = 0.54).
Two-Sample Mr Result
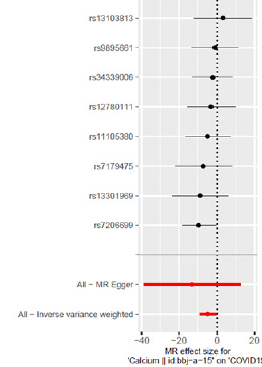
In Asian populations, serum calcium concentration is associated with a reduced risk of COVID-19 (Figure 1 & 2).
Heterogeneity Test Result
The Cochran’s Q test result of IVW is P=0.87, indicating that there is no heterogeneity in SNPs; the Cochran’s Q test result of MREgger regression is consistent with it.
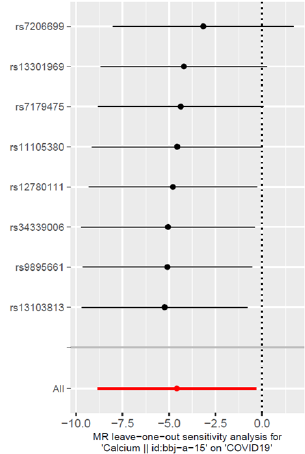
The “Leave-one-out” sensitivity analysis results of this study are shown in (Figure 3). No matter which SNP is eliminated, the analysis results of the remaining 7 SNPs are similar to the analysis results of all the included SNPs, and no influence on the estimated value of causal association is found. Larger SNPs (Figure 3).
Conclusion
Appropriate supplementation of serum calcium concentration can help prevent the onset of COVID-19.
Discussion
This project uses large-scale GWAS aggregated data and uses two-sample MR to explore the causal relationship between serum calcium concentration and the risk of COVID-19 in Asian populations. The results show that serum calcium can reduce the risk of COVID-19.
1. Previous studies have shown that in European populations, serum calcium concentration is negatively correlated with the risk of COVID-19. This research has reached unanimous conclusions. This result may be explained by the following two mechanisms: Calcium ions can regulate cell function and help regulate enzyme activity, thereby enhancing the body’s immune function;
2. Calcium ions can reduce the permeability of capillaries and cell membranes, Prevent oozing, inhibit inflammation and edema. In other words, the biological pathway involved in calcium ions may partly coincide with the pathogenesis of COVID-19.
This study has the following advantages: First, the use of public GWAS data can save research cost and time; Second, the two GWAS databases are both from Asian populations and are independent of each other, avoiding population bias; Third, it is not the same as using a single SNP as the IV In comparison, using 8 SNPs together as IVs can increase the proportion of calcium ion variation that can be explained by genetic variation; fourth, compared with traditional experimental studies, MR simulates a more realistic random allocation process, and the research design Relatively simple, research and implementation will not violate ethics. At the same time, this study also has some limitations. 1. [12] First of all, MR cannot further explore and explain the biological mechanism of the influence of genetic variation on the pathogenesis of COVID-19. Second, MR assumes that there is a linear relationship between exposure and outcome. If there is a non-linear relationship between calcium ion concentration and the risk of COVID-19, MR will not be suitable for causal inferences between the two. Third, this study uses two GWAS aggregated databases, lacking individual level data, unable to conduct subgroup analysis of age or gender, and unable to compare the differences in causal effects between subgroups. At the same time, the small sample size of the COVID-19 database will cause problems.
The statistical power of causal inference is reduced. Finally, this study did not adjust the immune-related indicators, and the MR method can only make preliminary inferences about its causality, and further research is needed to explore it. In summary, this study used two samples of MR to investigate the serum calcium concentration and the risk of COVID-19. The results showed that there is a negative causal relationship between the two, that is, the increase in serum calcium concentration can reduce the incidence of COVID-19. Risk, which can provide a certain reference value for the clinical treatment process.



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.