Surface Marker Analysis to Predict Successful Reprogramming to Pluripotency
Introduction
Induced pluripotent stem cells (iPSCs) are generated from somatic cells by introduction of Yamanaka factors comprised of Oct4, Sox2, Klf4, and c-Myc (OSKM), and they are highly desired for applications in regenerative medicine, tailor-made therapy, and disease modeling [1,2]. iPSCs derived from patients’ somatic cells are thought to minimize graft rejection when they are implanted into the target tissue. In addition, it is not necessary to consider an ethical issue accompanied with embryonic stem cells. However, due to the low efficiency of iPSC formation and heterogeneity in the reprogramming process, it remains difficult to gain insights into the molecular basis of successful reprogramming to iPSCs. To address these issues above, we have recently revealed that a certain surface marker profile determines a cell population in which bona fide reprogramming progenitors is highly enriched [3]. We previously performed microarray analysis to identify surface marker profiles for bona fide iPSC progenitors [3].
Cell Culture and iPSC Induction
MEFs used as somatic cells for this study were prepared from C57B/6 mouse embryos E13.5 to E14.5. MEFs were cultured in a medium containing Dulbecco’s Modified Eagle Medium (Nacalai Tesque, Japan), 10% fetal bovine serum (Nichirei Bioscience, Japan), 2 mM L-glutamine (GlutaMAX, GIBCO), and 1% (10,000 U/L and 10 mM) penicillin-streptomycin (Wako, Japan). We performed iPSC induction as previously described [3]. We used retroviral vectors as follows: pMXs-mOct4, pMXs-mSox2, pMXs-mKlf4, pMXs-mcMyc, pMXs-null. Following transfection of these plasmids to HEK293T packaging cells, the supernatant of the medium for transfected cells were used as virus-containing solution. The day when MEFs were incubated in the virus-containing solution was set as day 0. Two days after infection, cells being reprogrammed were maintained in a LIF-supplemented medium for undifferentiated iPSCs as previously described [3]. Cell culture was performed in the condition at 37°C and 5% CO2 in a humidified incubator.
Based on a standard protocol for ABC staining, immunolabeling of iPSC colonies were performed using the Vecta Stain ABC kit and ImmPACT DAB substrate (Vector Laboratories) with polyclonal rabbit anti-mouse Nanog (Calbiochem) antibodies as previously described [4]. Nanog-positive colonies were counted under a stereomicroscope.
Cells infected with OSKM were sorted by FACS (FACSAria III, BD Biosciences) at 2, 5, or 13 days after infection using FITC-conjugated or APC-conjugated anti-Sca1, PE-conjugated anti-CD34, PEconjugated anti-CD44, Alexa Fluor 647-conjugated anti-CD54, and PE or APC-conjugated anti-SSEA1 antibodies. After trypsinization, cultured cells were resuspended with antibody-containing D-PBS (-) containing 1% fetal bovine serum (Nichirei Bioscience) and 1 mM 2NA (EDTA-2Na) at 4°C for 1 hour, then washed twice. The cells were resuspended at up to 5.0 x 106 cells/mL in the buffer solution and subjected to FACS. Acquired data were analyzed with FlowJo software (FlowJo, LLC).
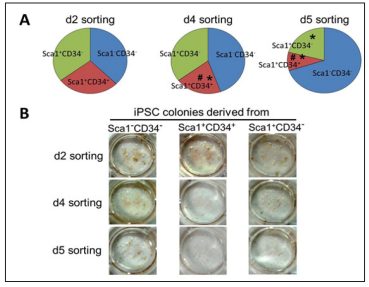
First, we examined the crucial timing to detect the Sca1-CD34- population for the most favorable efficiency of reprogramming to pluripotency. We performed FACS analysis after OSKM-infection in MEFs at days 2, 4, and 5. The sorted cells were subsequently replaced and cultured for iPSC generation. Subsequently, to evaluate reprogramming efficiency, we performed immunostaining using antibodies against Nanog, one of the specific markers for undifferentiated iPSCs. We observed significant increases in reprogramming efficiency originated from the Sca1-CD34- cell population sorted at day 4, compared to day2 at which there were almost no differences among all cell populations. However, the efficiency was remarkably increased in Sca1-CD34- cells sorted at day 5 (Figure 1A and 1B), demonstrating that detecting the Sca1-CD34- profile at day 5 is most effective to predict successful reprogramming to iPSCs.
a) And representative photographs
b) Of Nanog-positive iPSC colonies derived from Sca1-CD34-, Sca1+CD34+, and Sca1+CD34- cell populations sorted at days 2, 4, and 5.
* p < 0.05 vs Sca1-CD34-, # p < 0.05 vs Sca1+CD34-.
 Whereas our previous study revealed that Sca1-CD34- cells exhibit high reprogramming to iPSCs, several studies addressed that a surface marker profile of SSEA1+ or CD44-CD54+ can prospect successful reprogramming [5,6]. Stadtfeld and colleagues used doxycycline- inducible vectors to temporally express OSKM. They found that downregulation of Thy1, a surface marker for fibroblasts and other differentiated cells types [5], and subsequent upregulation of SSEA1 are the dynamic profile changes indicating an intermediate cell population with high potential to become iPSCs [6]. O’Malley et al. examined progression of the reprogramming process with expression profile changes of surface markers, CD44 and CD45. As a result, the CD44-CD54+ population sorted in the relatively late stage also displayed efficient iPSC colony formation to fully reprogrammed iPSCs [7].
Whereas our previous study revealed that Sca1-CD34- cells exhibit high reprogramming to iPSCs, several studies addressed that a surface marker profile of SSEA1+ or CD44-CD54+ can prospect successful reprogramming [5,6]. Stadtfeld and colleagues used doxycycline- inducible vectors to temporally express OSKM. They found that downregulation of Thy1, a surface marker for fibroblasts and other differentiated cells types [5], and subsequent upregulation of SSEA1 are the dynamic profile changes indicating an intermediate cell population with high potential to become iPSCs [6]. O’Malley et al. examined progression of the reprogramming process with expression profile changes of surface markers, CD44 and CD45. As a result, the CD44-CD54+ population sorted in the relatively late stage also displayed efficient iPSC colony formation to fully reprogrammed iPSCs [7].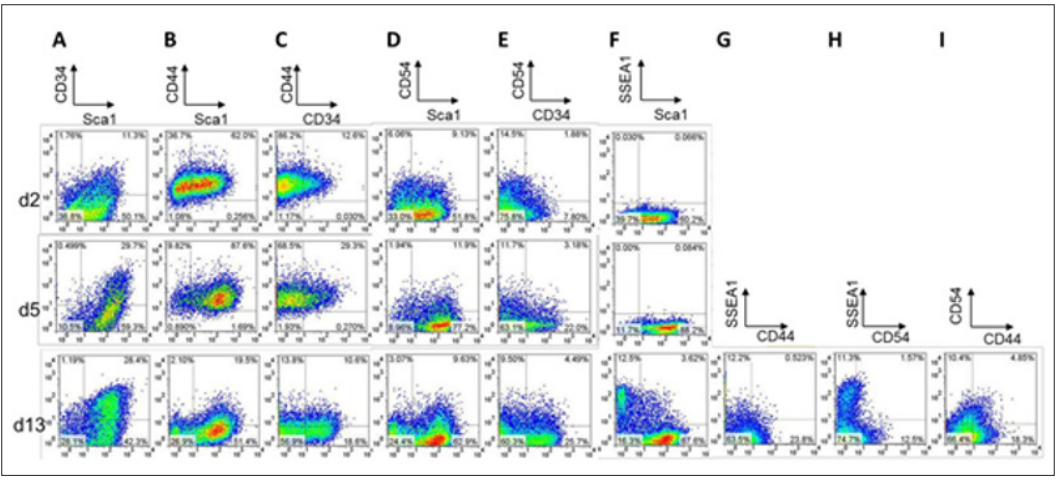 Figure 3: FACS plots of correlations between SSEA1 enrichment and various surface marker profiles in MEFs undergoing reprogramming to pluripotency at day 13 after OSKM-infection. (A) and (B) Correlation between SSEA1 enrichment and Sca1/ CD34 profiles (A) and CD44/CD54 profiles (B) are shown.
Figure 3: FACS plots of correlations between SSEA1 enrichment and various surface marker profiles in MEFs undergoing reprogramming to pluripotency at day 13 after OSKM-infection. (A) and (B) Correlation between SSEA1 enrichment and Sca1/ CD34 profiles (A) and CD44/CD54 profiles (B) are shown.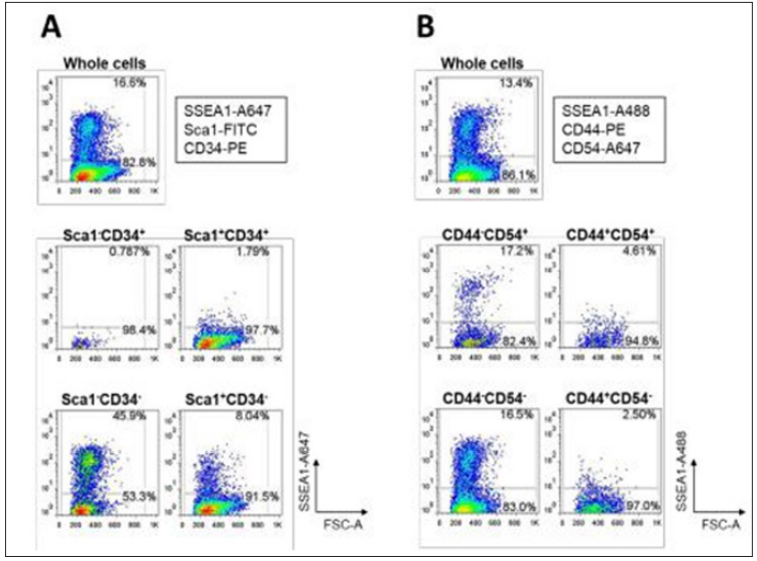 These results also support our proposal that reprogramming behavior of Sca1-CD34- cells seems to overlap with that of SSEA1-positive cells. Different characteristics of the CD44-CD54+ population compared to those of the Sca1-CD34- population might be due to different detection systems for undifferentiated iPSCs. O’Malley and colleagues used Nanog-eGFP reporter cells in which GFP is expressed under the control of the Nanog promoter [7], whereas we detected undifferentiated iPSCs with immunocytochemistry using anti-Nanog antibodies.
These results also support our proposal that reprogramming behavior of Sca1-CD34- cells seems to overlap with that of SSEA1-positive cells. Different characteristics of the CD44-CD54+ population compared to those of the Sca1-CD34- population might be due to different detection systems for undifferentiated iPSCs. O’Malley and colleagues used Nanog-eGFP reporter cells in which GFP is expressed under the control of the Nanog promoter [7], whereas we detected undifferentiated iPSCs with immunocytochemistry using anti-Nanog antibodies.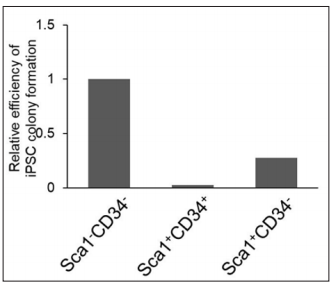 Conclusion
ConclusionSca1-CD34- is the surface marker profile detectable at the early reprogramming stage, and it reflects successfully reprogrammed cells, half of which exhibit SSEA1 expression at the late stage. The surface marker combination of Sca1 and CD34 is a more sensitive predictor for bona fide iPSC progenitors in the early phase, and it will be useful to shed light on the molecular mechanism of successful reprogramming to iPSCs.
More BJSTR Articles : https://biomedres01.blogspot.com



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.