Genetic Research in Axial Spondyloarthropathies - What Do We Know?
Introduction
Spondyloarthropathies (SpAs) are a group of diseases that are characterized by arthritis in the spine and peripheral joints. Apart from ailments from the osteoarticular system, there are also nonarticular symptoms, including inflammation of the middle layer of the eye between the retina and the sclera (uveitis), psoriasis and inflammatory bowel diseases. The onset of the disease is difficult to observe. Due to the lack of a pathognomonic test, diagnosis is based on a combination of physical examination, laboratory test and imaging results. The pathogenesis of SpAs is not fully understood, and genetic, environmental and immunological factors are assumed to play a role [1]. Despite numerous studies, we are not able to predict the course of the disease or the occurrence of extra-articular symptoms. The lack or loss of subsequent drug effectiveness observed in some patients is also a problem. Axial SpA (axSpA) usually starts before 45 years of age. Progressive disease leads to restricted mobility, resulting in difficulties in daily functioning of the patient or even disability. AxSpA is associated with a burden in terms of physical function, mood disturbance, work impact and quality of life impairment [2]. It was recognized that there is often a period where classic signs and symptoms of axial inflammatory disease are present in the absence of radiographic changes in the sacroiliac joints fulfilling the criteria for ankylosing spondylitis (AS); this period was subsequently given the name nonradiographic axSpA [3].
The average delay between the onset of symptoms and the diagnosis of axSpA is estimated to be 5 to 7 years in the United States. Several factors may contribute to the delay in diagnosis, including the high prevalence of back pain (most commonly due to mechanical aetiologies) in the general population. The lack of specific physical examination findings in patients with early axSpA and the absence of extra-spinal manifestations have been reported to impair early diagnosis. The lack of biomarkers unique to axSpA, younger age at onset, and gradual disease onset may also contribute to delayed referral for evaluation by a rheumatologist [4]. The first-line drugs are nonsteroidal anti-inflammatory drugs (NSAIDs). In the case of long-lasting high-activity disease, continuous use of NSAIDs is recommended, and in the case of stable disease, NSAID use is recommended, if necessary, as when pain occurs (an on-demand strategy). In patients with peripheral arthritis, the inclusion of sulfasalazine may be considered. Glucocorticosteroids are acceptable but only for joint injections. In the second stage, biological disease-modifying antirheumatic drugs (bDMARDs) are used if standard treatment is not effective. An important role is played by rehabilitation, which prevents the stiffening of spinal column tissues and peripheral joints. Unfortunately, treatment is problematic in some patients. Many patients cannot take medications due to increased risks for adverse events. Approximately one-third of patients treated with anti-TNFs will have an inadequate response or lose responsiveness to these drugs over time, and in many patients, this may be the result of the development of antidrug antibodies [5].
A key clinical question in AS is whether to start treatment with a TNF inhibitor (infliximab, etanercept, adalimumab, certolizumab, golimumab, and their biosimilars), an IL-17 inhibitor (secukinumab and ixekizumab), or a targeted synthetic DMARD (such as tofacitinib). Another question is when to discontinue therapy [6,7]. The purpose of this study was to describe the genetic basis of axSpA and to present potential genetic markers of severe disease, extraarticular symptoms and response to treatment. We focused on the most common SpA – AS.
Pathogenesis
The pathogenesis of AS is not fully understood. The role of genetic factors, intestinal/skin barrier disorders, and infectious factors, which, with the participation of environmental factors (mechanical stress), lead to the development of inflammation, is emphasized. Among immunological disorders associated with pathogenesis, the most interesting are two pathways in the inflammatory responses, probably located at the end of the immune response hierarchy, the tumour necrosis factor alpha axis (TNFα) and the interleukin-23/ interleukin-17 axis (IL-23/IL-17). How the TNFα and IL-17 pathways are connected and whether there is a hierarchical order between the two are not clear [8]. Further understanding of the cellular and molecular regulatory mechanisms of the IL-23/IL-17 axis and other inflammatory cytokines may provide a promising strategy in SpA treatment [9].
The Role of Genetics in AS Pathogenesis
Accumulating evidence has suggested that AS is highly heritable. Human leukocyte antigen (HLA)-B27 is one genetic factor with a convincing association with AS, and HLA-B27 was reported to be present in 94,3% of patients. However, twin and family studies suggest that HLA-B27 can explain only less than 30% of the overall risk for AS, meaning that there are other genes related to the genetic disorder of AS. Several theories have been proposed explaining the role of HLA-B27 in the pathogenesis of axSpA. The three most prominent, not mutually exclusive theories are the “arthritogenic peptide hypothesis”, “the heavy chain homodimer hypothesis” and the “HLA-B27 misfolding hypothesis”. As a result of HLA-B27 molecule interactions with leukocytes or by inducing cellular stress, autoimmune processes are activated, including the IL-23/IL-17 pathway. Recently, scholars have also aimed to investigate other inflammatory biomarkers for AS, including interleukin IL-8, TNFα, C-reactive protein (hsCRP) and C-C motif chemokine 11 (CCL11), but studies that have focused on genetic biomarkers are limited [10,11]. Nevertheless, more than 100 nonmajor histocompatibility complex (MHC) loci have been identified at genome-wide significance levels, either in studies of AS alone or in subset-based meta-analysis of related diseases. This level of significance is considered robust, and most of the loci have crosssupport between studies. The loci can be divided into the following categories: cytokines and cytokine receptors, mucosal immunity factors, M1-aminopeptidases, transcription factors and intergenic regions. Considering cytokines and cytokine factors, these loci can be divided largely into either IL-23 pathway or TNF pathway genes. There is a lack of large-scale pharmacogenomic studies in AS [12].
HLA-B27 Genetic Diversity and Its Relationship to AS
HLA represent a group of highly polymorphic genes that reside in the major MHC which is located within the 6p21.3 region on the short arm of chromosome 6 and encodes many of the proteins of the immune system. These include HLA-class I genes that are codominantly expressed on the cell surface presenting intracellularly derived peptides to CD8 positive T cells. HLA-B27 belongs to a family of closely related cell surface proteins encoded in the HLA-B locus. Many of the mutations are located within introns and thus are silent or occur in exons but do not cause amino acid changes. Therefore, at the translated protein level, there are over 150 known subtypes of HLA-B27 based on one or more amino acid sequence differences [13]. Sometimes the differences may concern only 1 amino acid, e.g., HLA-B27:04 varies from HLA-B27:05 only at position 152 (Val to Glu) [14]. Different subtypes of HLA-B27 are distributed unevenly worldwide and with different strengths of association with AS disease. The most frequent subtype is HLA-B27:05, which is found in all races and ethnicities. Data about the association of HLA-B27 subtypes with disease risk suggest that HLA-B27:05, HLA-B27:04 and HLA-B27:02 are strongly associated, but HLA-B27:06 and HLA-B27:09 are not (or weakly) associated, with AS. The relationship between HLA-B27 polymorphisms and the clinical characteristics of AS patients has also been demonstrated by some investigations, but the reports are conflicting [14]. On the other hand, it is worth highlighting that HLA-B27 has a protective role in HIV and HCV infections. The association among HLA-B27 homozygosity, AS risk and its clinical characteristics has also been investigated. Homozygosity increases AS risk but does not affect clinical symptoms [14]. Recently, Wu et al. concluded that HLA-B27 heterozygotes (HLA-B27/B46) had more peripheral joint involvement among all HLA-B27(+) AS Chinese patients [15].
AS – A Hereditary Disease
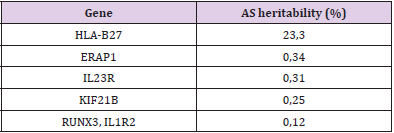
It has long been known that AS runs strongly in families, with the risk of disease in first-degree relatives of AS patients being >52 times that of unrelated subjects. The recurrence risk for AS in monozygotic twins is 63%, in first-degree relatives is 8,2% and in second-degree relatives is 1,0%. The parent–child recurrence risk is 7,9%, and the sibling–sibling recurrence risk is 8,2%. HLAB27- positive first-degree relatives of AS patients are 5,6–16 times more likely to develop disease themselves than HLA-B27-positive carriers in the general community [16]. Even though HLA-B27 plays an undisputedly critical role in disease pathogenesis, estimates suggest that it accounts for only 20–25% of the total heritability and 40% of the genetic risk. Fewer than 5% of HLA-B27 carriers in the general population develop disease. Each of the non-HLA-B27 gene SNPs individually confers a small amount of risk, with odds ratios ≤1,65 [17] (Table 1). All non-MHC loci contribute another ~10% of AS heritability [18]. The question arises – What about the rest? To summarize, in AS, only up to 30% of the heritability has been elucidated. One reason for this shortfall is the requirement for large sample sizes (in the tens of thousands or higher) for the discovery of genes that have a small impact on overall susceptibility (associations with disease with an odds ratio of 1,1 or less). Other sources of genetic contribution are rare variants, the discovery of which will require extensive resequencing studies. Small gene copy number variants (CNVs) and insertions/deletions are extremely difficult to genotype using current high-throughput array technology (which is optimized for SNP genotyping) and thus remain a potential source of missing heritability. Epigenetic factors, such as differences in methylation patterns, might also have a role in conferring susceptibility, but the heritability of such influences is minor.
Finally, epistasis (gene–gene interaction) is an area of recent investigation. Heritability estimates such as the one mentioned above are calculated from models of pathogenesis that allow for only additive effects, that is, in the absence of gene–gene interactions. Validation studies that use multi-marker, as opposed to single-marker, analyses in independent cohorts are reported to have an improved capacity for risk prediction; such studies are the focus of ongoing investigations as the statistical methodologies are being developed and refined [19]. (Table 1) Genes with the greatest contribution to AS heritability [20].
Non-MHC Genetic Polymorphisms Related to AS
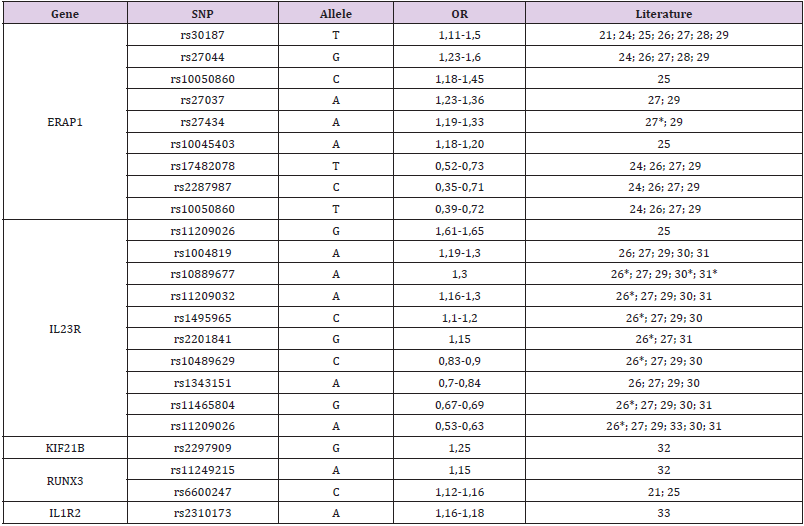
The discovery of AS-related genes provides insight into disease pathogenesis and immune system function. The evidence confirms that aberrant peptide processing before MHC class I presentation and alterations of the IL-23 pathway are key elements in the pathogenesis of AS. It is worth noting that some loci associated with AS overlap with other immune-mediated diseases, such as inflammatory bowel disease, rheumatoid arthritis or psoriasis [21]. Some of them may appear as extra-articular symptoms of AS. Some of the most notable genetic findings from studies involving AS are the discoveries implicating the involvement of aminopeptidases – endoplasmic reticulum aminopeptidase 1 (ERAP1), ERAP2, and leucyl-cysteinyl aminopeptidase (LNPEP) – and genes in both the TNF and IL-23 pathways. ERAP1 has the second strongest association with AS and displays a synergistic interaction with HLA-B27. This association is lost in HLA-B27-negative patients, but a weaker association with ERAP2 can be seen in both HLAB27- negative and HLA-B27-positive patients. ERAP1 and other aminopeptidases appear to play a significant role in trimming peptides transported from the cytosol to the endoplasmic reticulum to optimal length (8 or 9 amino acids) for loading on HLA class I molecules [22,23]. There are many papers about genes associated with AS susceptibility. Single-nucleotide polymorphisms (SNPs) of the most important genes and their combined odds ratios (ORs) are shown in (Table 2). Most important SNPs of non-MHC genes with the highest AS heritability.
Table 2: Most important SNPs of non-MHC genes with the highest AS heritability.
Note: OR (odds ratio) from the literature, only when it was statistically significant.
If present, the OR for all ethnicities was preferred. A single OR was enrolled when only one ethnic group had a statistically significant OR (e.g., European).
* - p statistically insignificant in this study
Evidence for a genetic relationship with disease activity seems to be limited. The association among AS disease activity, function, spinal mobility and IL-17 or IL-23 is not fully understood. The dysregulation of this pathway may lead to systemic chronic autoimmune inflammation, causing extra-articular involvement [24]. Moreover, İnal et al. suggested that the IL-17F polymorphism may be associated with susceptibility to AS, disease activity and functional status in Turkish patients [25]. Another study proposed IL-12B, IL-6R, RANKL, STAT4 and FCRL4 gene polymorphisms as promising biomarkers for diagnosis and prognosis in AS patients [26-40]. Other genes associated with disease activity are IL17RA and JMY and region 2p15 [41-43]. It is worth noting that many AS patients with high disease activity often do not show corresponding high CRP levels. An explanation for this might be a genetic contribution to variation in CRP levels. This observation may be important for the interpretation of disease activity scores such as the ASDAS, on which clinical decisions regarding drug selection are based [44]. Further research showed that the CRP rs3091244 SNP was associated with an increased risk of AS. Moreover, it could serve as a biomarker for a good response to etanercept treatment in AS [45]. More studies are needed in a larger group of patients to confirm these results. Another important topic related to axSpA is the occurrence of extra-articular manifestations. The most common is anterior uveitis, which affects 25% to 35% of patients.
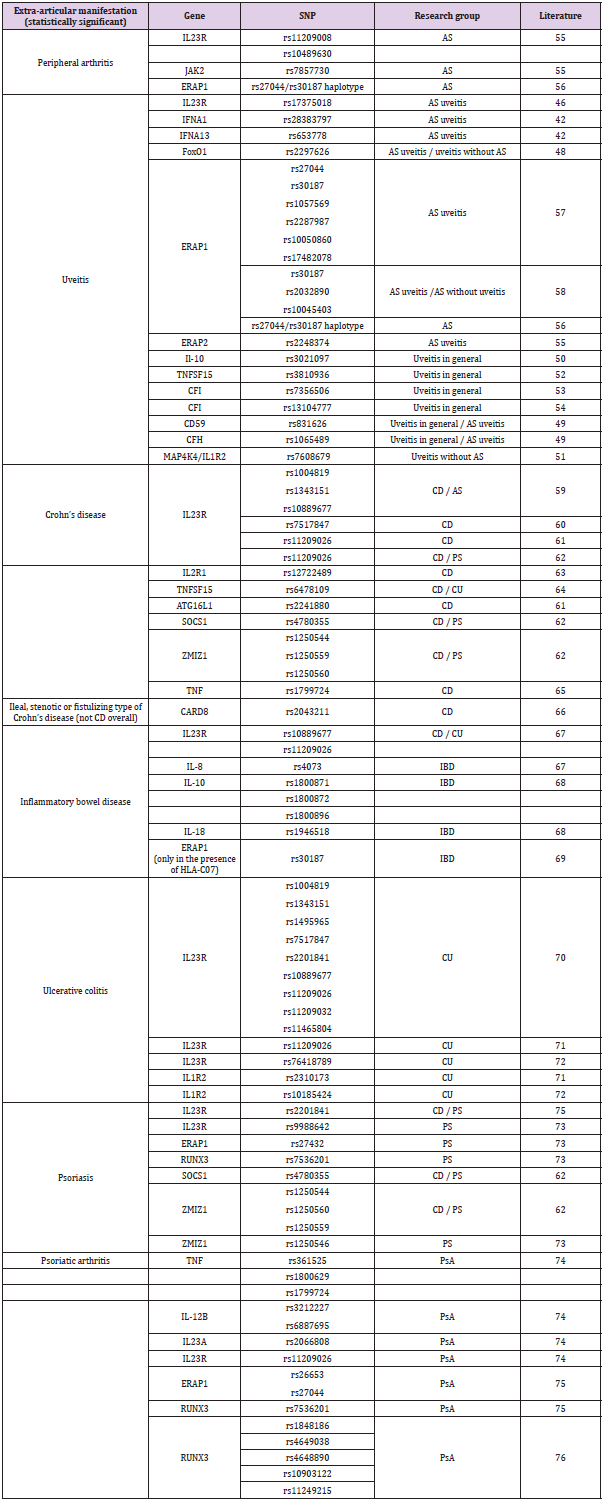
Studies conducted mainly in China have shown an association of IL23R, FoxO1, IFNA1, IFNA13, and CFH gene polymorphisms with the occurrence of uveitis in patients with AS [46-49]. Other genes with a confirmed association with uveitis in AS patients are ERAP1 and ERAP2. Research should be extended to include polymorphisms of other uveitis-related genes to include AS patients, e.g., IL- 10, MAP4K4/IL1R2, TNFSF15, CFI, CD59, and CFH [49-54]. The influence could be related to sex, the presence of HLA-B27 or AS status. SNPs of genes associated with peripheral arthritis and extraarticular symptoms in AS are shown in Table 3. We added psoriatic arthritis to show possible similarity to another SpA. (Table 3) SNPs of genes associated with peripheral arthritis and extra-articular symptoms in AS. As we mentioned above, treatment is not effective in every patient. Is it possible to find the right drug for a specific patient to improve efficacy or reduce side effects? [55-60]. The answer to this question can be found in genetic SNPs, resulting in the formation of proteins with different activities.
Table 3: SNPs of genes associated with peripheral arthritis and extra-articular symptoms in AS.
Note: CD - Crohn’s disease
UC - Ulcerative colitis
PS - Psoriasis
PsA - Psoriatic arthritis
IBD - Inflammatory bowel disease (CD and UC)
Pharmacogenetics and pharmacogenomics have confirmed that genetic polymorphisms may have an impact on drug metabolism, drug targets, or drug receptors, resulting in interindividual variability in drug disposition and efficacy. Studies have demonstrated that variants in cytochrome P-450 (CYP) genes can result in differences in the expression and function of their relevant encoding enzymes, thus affecting the patient’s response to drugs. A Chinese study indicated the effect of CYP2D6*10 and CYP3A5*3 polymorphisms on the efficacy of anti-TNF etanercept treatment for AS patients [61]. Anti-TNFα agents have been proven highly effective in a large number of patients, but the early identification of patients more prone to show an optimal and stable response in the long term remains an open issue. What about the TNFα gene itself? Scientists identified the TNFα rs1800629 and IL-6 rs1800795 promoter polymorphisms as useful genetic biomarkers of response to TNFα blockers in a multicentre retrospective cohort of patients with SpA by considering, as the primary outcome, the long-term retention rate for treatment with the first TNFα blocker [62]. A better response to anti-TNFα treatment was also confirmed in the group of RA, PsA or AS patients [63].
Unfortunately, study results are not consistent. The contradictory data concern rs1800629; in China, no such relationship between rs1800629 and response to anti-TNFα treatment has been confirmed, in contrast to Europe [64]. Nevertheless, the results from a meta-analysis of papers from all around the world indicate that TNFα rs1800629, apart from rs361525, could predict the response to etanercept much more powerfully than the response to infliximab/adalimumab [65]. In the following year, a relationship regarding the effectiveness of anti-TNF treatment and rs1800629 was also confirmed in Asia. The study group consisted of SpA and inflammatory bowel disease patients [66]. Moreover, in the Bulgarian population, TNFα rs1800629 was found to be associated with genetic susceptibility to AS, age at onset and disease severity [67]. AS severity dependence on the TNFα gene has been confirmed in Norway, as a reduced risk of uveitis and better spinal function [68].
The influence of tumour necrosis factor receptor 1A (TNFRSF1A) and TNFRSF1B gene polymorphisms also seems to be interesting for medicine. In a study lasting 12 months, an association with the long-term therapeutic efficacy of etanercept was confirmed for AS (rs1061622). Additional data indicated dependence on AS susceptibility (rs767455) and severity measured as chest expansion (rs1061622) [69]. In Europe, researchers evaluated various TNF inhibitors, such as infliximab, adalimumab, etanercept, and golimumab, and found a different polymorphism in the TNFRSF1A gene (rs1800693) that impacted the response to anti-TNF therapy for SpA [70]. Another genetic marker of etanercept therapy may be the ABCB1 gene [71]. Schiotis et al. searched for pharmacogenomic markers responsible for nonresponse to anti-TNFα agents in previously untreated AS patients. They found an association of nonresponse to anti-TNFα agents with the MIF gene rs755622, IL18RAP gene rs917997, TNFRSF1B gene rs1061622, ARFGAP2 gene rs3740691 and IL-10 gene rs1800896 polymorphisms. The strongest predictor of nonresponse to anti-TNFα agents was the IL18RAP gene. Using a candidate SNP approach, they developed a genetic model of nonresponse. The validation of this genetic model in prospective studies may lead to the design of a clinico-genetic algorithm to initiate biological treatment [72].
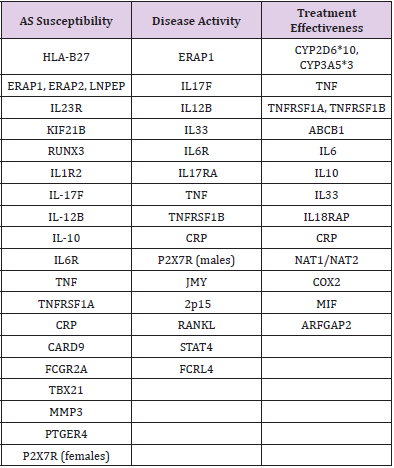
Table 4: Genes and their relationship with AS susceptibility, disease activity and treatment effectiveness.
Note: [Based on 20, 21, 22, 23, 28, 29, 31, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 56, 77, 79, 80, 81, 82, 83, 84, 85, 87, 88, 89, 90, 91, 92, 93, 94].
Recently growing body of evidence highlights the role of the IL-33 signaling pathway in inflammatory arthritis, such as AS. This cytokine functions as an alarmin, alerting the immune system and triggering the inflammatory process. In the Caucasian population, a significant association in the IL-33 rs16924159 genotype distribution with regard to disease activity and anti-TNF therapy efficacy was found (89). The effect of classic DMARDs such as sulfasalazine or NSAIDs can also be predicted using genetic markers such as NAT1/NAT2 and COX2, respectively [73-75]. The development of biological drugs acting on various inflammatory cytokines raises the following question: what kind of therapy should be used in a particular patient? What is better: anti-TNF or anti-IL-23/IL-17 pathway drugs? Based on such genetic profiles, clinicians can recommend appropriate treatment to patients. Genetic research should be conducted in different populations. The association of gene polymorphisms with AS can be worldwide or can concern only one race. For example, ERAP1 rs27044 appeared to be significantly correlated with AS in both Asians and Caucasians. For ERAP1 rs30187, the findings of genotypic comparisons supported that the association existed only in Caucasians but not Asians [76-82]. Stratification by ethnicity identified a significant association between some SNPs of IL-23R and AS susceptibility in Europeans and Americans but not in Asians [83-94]. The authors of many studies emphasize the need for further research in other countries. (Table 4) shows the most important genes and their association with AS. (Table 4) Genes and their relationship with AS susceptibility, disease activity and treatment effectiveness.
Conclusion
AxSpA affects young and economically and socially active people. We are currently unable to predict the course of disease or the presence of extra-articular symptoms. Which patients are at risk of complications? Which patients are at risk of adverse reactions to treatment? Due to the risk of disability, it is advisable to include appropriate treatment quickly. Genetic markers could improve the diagnosis of this disease. Early identification of patients at risk of a severe course of the disease or extra-articular symptom occurrence would enable early intensification of the therapy. In addition, understanding the expression of genes involved in the pathogenesis of the disease will allow new types of drugs to be developed e.g. maybe the IL-33 blockers will be effective? Currently, new biological drugs are being developed. These drugs act on different factors within the inflammatory pathway. Which treatment line should be chosen first? In the case of ineffectiveness, which drug should be chosen as the second- and third-line therapies? Which patients will be non-responders? There are many questions concerning therapy. The real revolution in medicine is yet to come. Scientific studies of many genes, their haplotypes and mutual interactions may allow the creation of a genetic model that predicts disease activity, the risk of extra-articular symptom occurrence or treatment response status with increasing effectiveness and minimizing side effects.
Such studies need to be carried out in different regions of the world to exclude racial differences. Patient genetic profile determination will allow new guidelines to be developed. Knowledge regarding the genetic markers of potential ineffectiveness of therapy and the risk of side effects may be key factors in the process of choosing the right medicine, thus personalizing treatment. Gene SNPs have the potential to be genetic markers of the effectiveness of therapy. This will result in better and faster treatment outcomes and will allow patients to remain physically active, improve patient quality of life or even extend patient lifespans The future is promising.
For more Articles on: https://biomedres01.blogspot.com/



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.