Difference in Leukemia Microenvironment Induced by PAX5 Mutations in Acute Lymphoblastic Leukemia
Background
Acute lymphoblastic leukemia (ALL) comes from a abnormal clonal expansion of immature lymphatic progenitors of B cell or T cell. ALL ordinarily violate bone marrow (BM), peripheral blood, and extramedullary sites. ALL accounts for a high proportion of pediatric leukemia and is common leukemia in adults. The prognosis of adult ALL is poor [1]. The outcome of pediatric ALL was obviously better than that of adult ALL, despite this, 10% of pediatric ALL relapse [1]. PAX5 (paired box domain gene 5) is one of the important genes which encodes a paired box domain transcription factor. PAX5 is essential for B-cell differentiation. PAX5 not only activates genes necessary for B cell differentiation, but also inhibits genes associated with lineage transformation [2]. PAX5 mutations were detected often in both childhood and adult B-ALL [3-5]. PAX5 mutations are an independent poor prognostic factor for disease-free survival in B-ALL children [6]. So PAX5 mutation plays an important role in the onset and recurrence of leukemia. The tumor microenvironment obviously influences therapeutic effect and clinical outcome. Tumor immune escape and angiogenesis takes part in tumorigenesis and relapse of ALL. T lymphocytes within the tumor microenvironment are preferred immune cells for targeting cancer. In addition to T cells, other immune cells also play a crucial part in leukemia immunity. Various cell populations shape the tumor microenvironment and mediate tumor immune escape through multiple interactions [7].
Except T cells, macrophages have become one of the most comprehensively investigated immune cell types within the tumor immune microenvironment. There is dual role of macrophage in terms of tumor immunity and the dual role of macrophage depends on the phenotype and metabolism of macrophage [8]. Except immune cells and tumor cells, tumor microenvironment consisted of endothelial cells, fibroblasts, extracellular matrix, cytokines. Endothelial cells are not only involved in angiogenesis, but also involved in tumor immune escape [9]. Among tumor cells, immune cells and stroma, there are intense communications via cell–cell contact-dependent mechanisms and/or soluble messengers [9]. Different mutations of tumor cells lead to different ways of immune escape. Mutations of P53 induced low T-cell infiltration [10] and angiogenesis [11] within the tumor. In previous research, PAX5 haploinsufficiency promoting leukemia may be related to immune tolerance [12]. Recent report suggested that PAX5 mutations took part in B-ALL relapse after treatment using anti CD19 chimeric antigen receptor (CAR) T -cell [13,14]. The understanding of PAX5 alterations-inducing changes in leukemia microenvironment is beneficial to understand the role of PAX5 alterations in the onset and recurrence of leukemia. In our research, we used bioinformatics analysis methods to estimate divergence of immune cells and to characterize differentially expressed genes (DEGs) between ALL with PAX5 mutations and ALL without PAX5 mutations in leukemia microenvironment.
Materials and Methods
Data Preparation
The mRNA expression profiling of B-ALL patients was acquired from Gene Expression Omnibus (GEO http://www.ncbi.nlm.nih.gov/geo, accession number: GSE11877). In the patients analyzed, there were 134 patients that underwent PAX5 mutations test. 70 of the 134 patients tested were positive for negative, and 64 of the 134 patients tested were negative for PAX5 mutations (Supplementary material Tab 1) [15-17].
Analysis of Differentially Expressed Genes (DEGs)
After the gene expression data were converted to standard annotation files, identification of DEGs was carried out using the LIMMA package of R [18]. The DEGs between ALL with PAX5 mutations and ALL without PAX5 mutations were identified using the standard of P value ≤ 0.01.
Evaluation of Immune Cell Ratio
CIBERSORT, as an online software, can evaluate the cell constituent of tissues based on normalized gene expression profiles (GEPs). We used this algorithm to evaluate the relative components of immune cell types of bone marrow of B-ALL [19]. Simply, the gene expression profiles were converted to standard annotation files and were submitted to the CIBERSORT web portal (http://cibersort.stanford.edu/). 22 immune cell types were set as the signature gene file. The analysis was conducted with 1,000 permutations. The immune cell types in bone marrow included T cells (Tfh cells, resting memory CD4+ T cells, activated memory CD4+ T cells, γδ T cells, CD8+ T cells, Tregs, naïve CD4+ T cells), B cells (memory B cells, naïve B cells, plasma cells), NK cells (activated NK cells and resting NK cells), macrophages (M0, M1, M2), activated and resting mast cells, activated and resting dendritic cells, neutrophils, monocytes, and eosinophils.
Functional Annotation Clustering of Degs by the DAVID
The DAVID (Database for Annotation, Visualization and Integrated Discovery) contains an integrated biological knowledge base and analytic tools, for extracting biological meaning from large gene lists [20]. The GO analyses of DEGs in the two clusters of the highest enrichment score were performed by the DAVID online database with P<0.05 set as statistically significant.
Construction of Protein Protein Interaction Network (PPI Network) and Co-Expression Network
The online database STRING (Search Tool for the Retrieval of Interacting Genes) offers particularly wide coverage and ease of access to protein interaction information [21]. The interactions between DEGs in the two clusters of the highest enrichment score using DAVID were derived based on STRING and the associations with a correlation coefficient > 0.4 were recognized as PPIs. These hub genes were strongly correlated with other genes (connected with >5 proteins). Coexpedia is a database of context-associated coexpression networks inferred from individual series of microarray samples of human and mouse of GEO. The co-expression network analyzed by Coexpedia (http://www.coexpedia.org/) [22]. Genes with score ≥2 were selected as hub genes.
Result
Identification of Degs Between all with Pax5 Mutations and Without Pax5 Mutations
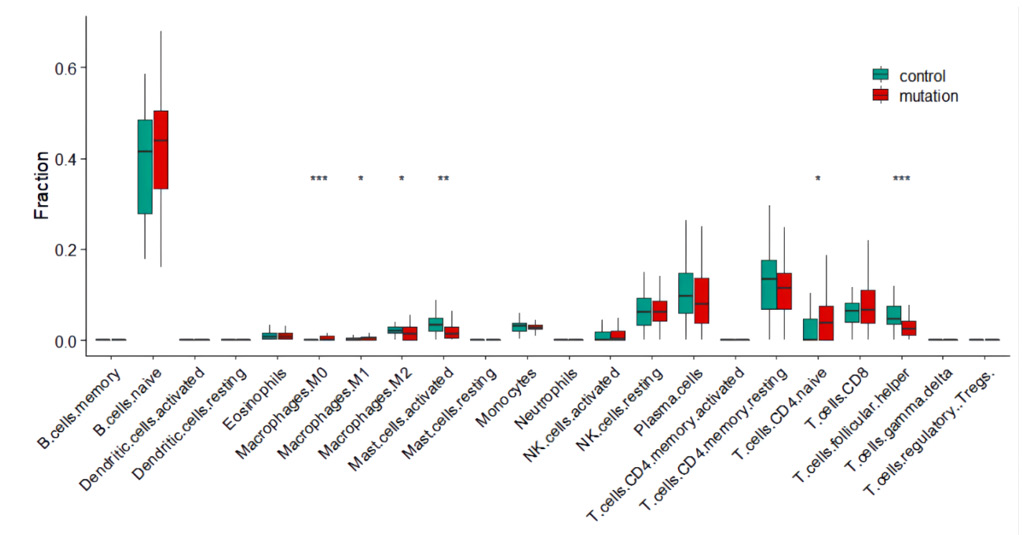
Based on the screening criteria (p-value of <0.01), there were 352 DEGs. There were 199 up-regulated DEGs and 153 downregulated DEGs (Supplementary material Tab 2). The estimation of bone marrow immune cell types of B-ALL. We first estimated the proportion of bone marrow immune cell types of B-ALL by the CIBERSORT software. The ratio of various immune cells was shown in (Figure 1). There were not statistically difference in the percentages of bone marrow T cells subtypes except follicular helper (Tfh) cells between PAX5 mutations B-ALL and wild type B-ALL. Apart from T cell subgroups, the proportions of macrophages and activated mast cells had obvious differences between two groups. The percentages of M0 cells in PAX5 mutations B-ALL was high and had obvious differences compared with PAX5 wild type B-ALL (p<0.001).
Figure 1: The compositions of immune cell subsets in bone marrow of B-ALL.
Note: The violin plot showed the percentage difference of immune cell subsets between B-ALL with PAX5 mutations(n=70) and B-ALL without PAX5 mutations(n=64).
*Means p < 0.05; ** means p < 0.01; *** means p < 0.001.
Functional Annotation Clustering of Degs and go Analysis
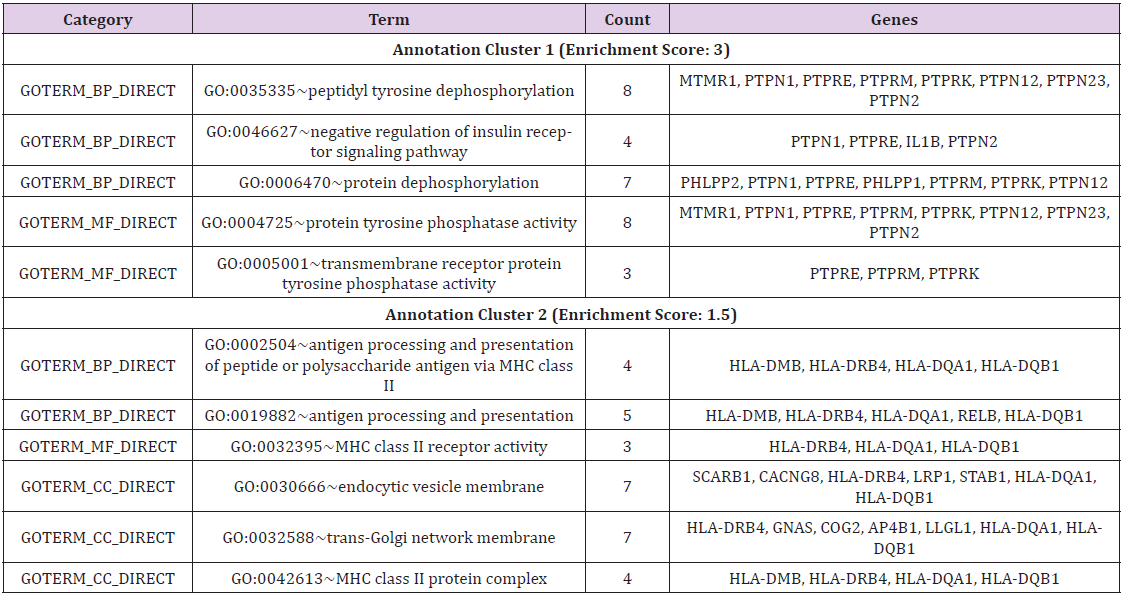
352 DEGs were clustered for functional annotation using DAVID, there were 53 annotation clusters (Supplementary material Tab 3). In order to better understanding functions of the DEGs, the GO biological process (BP), GO cellular component (CC), GO molecular function (MF) analyses were performed in the two clusters of the highest enrichment score(p<0.05). GO analysis hinted that the DEGs analyzed between PAX5 mutations ALL and wild type ALL were classified in 5 significant BP categories, 3 significant MF categories and 3 significant CC categories (Table 1). From BP analyses, most were clustered in antigen presentation including antigen presentation via MHC class II and protein dephosphorylation, especially tyrosine dephosphorylation. From CC analyses, most were clustered in MHC class II protein complex, trans-Golgi network membrane and endocytic vesicle membrane which were necessary for biological processes of antigen presentation [23]. From MF analyses, most were clustered in MHC class II receptor activity, protein tyrosine phosphatases (PTPs) activity which were required for extensive biological processes including antigen presentation [24]. GO analysis showed that most of these DEGs were MHC class II genes and PTPs genes, which reflected antigen presentation differences in biological processes, cellular component and molecular function. These DEGs may be involved in changes in macrophages.
Table 1: Functional classification of DEGs in the two clusters of the highest enrichment score(p<0.05).
Note: GOTERM_BP_DIRECT: GO biological process categories, GOTERM_CC_DIRECT: GO cellular component categories, GOTERM_MF_DIRECT: GO molecular function categories
Construction of PPI Network and Co -Expression Network
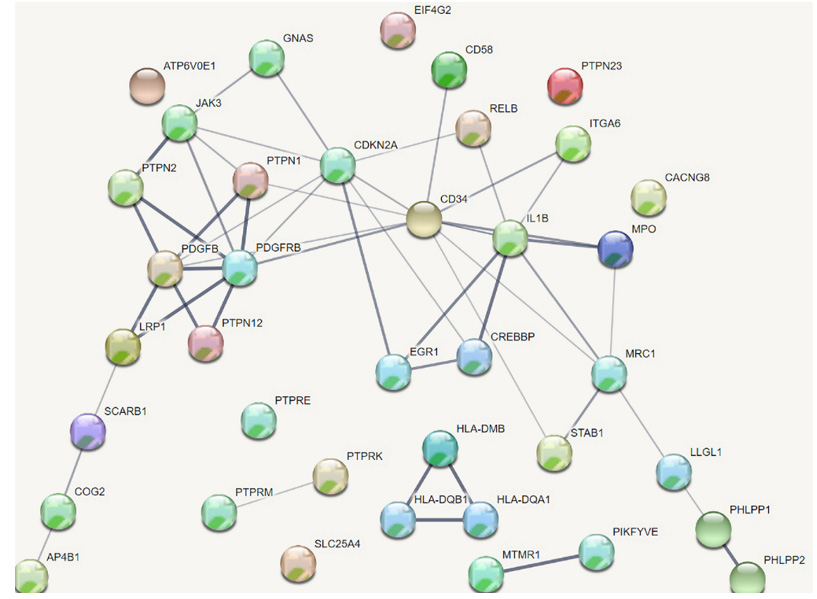
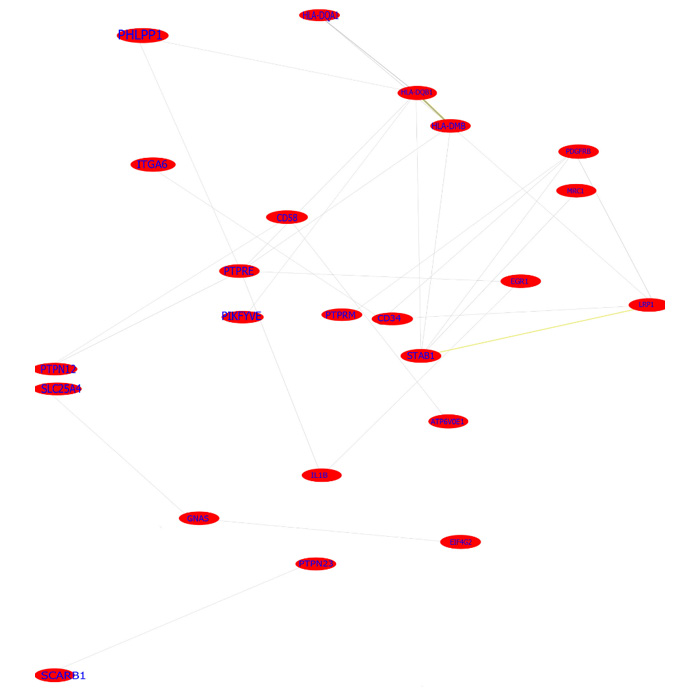
39 DEGs in the two clusters of the highest enrichment score using DAVID were included in the analysis of PPI network and co -expression network. The analysis of PPI networks found that there were 32 genes which were related to each other. The total DEGs PPI network contained 38 nodes and 50 edges (interactions) (Figure 2). Six genes—IL1B, CD34, CDKN2A, JAK3, PDGFRB and PDGFB— were strongly correlated with other proteins (connected with >5 proteins). The analysis of co-expression networks showed that 16 DEGs with score ≥2 was selected as hub genes. The co-expression network analyzed by Co expedia is shown in (Figure 3), presenting the co-expression relationships between hub genes. Three genes— IL1B, CD34, PDGFRB were common hub genes from the analysis of PPI network and co -expression network. These hub genes were marker genes of angiogenesis and played a crucial role in interactions between angiogenesis and macrophages.
Discussion
Tumor microenvironment takes part in leukemogenesis and relapse of ALL. In tumor microenvironment, immunologic escape and angiogenesis are important factors affecting clinical outcomes. Besides T cells, all kinds of immune cells and stroma within the tumor microenvironment also play the key role in tumor immune tolerance. In ALL leukemic microenvironment, there might be situations of high prevalence of regulatory T cells (Tregs), which inhibited innate and acquired immune responses [25,26]. Granulocytic myeloid-derived suppressor cells(G-MDSCs) participated in escaping immune surveillance in B-ALL, and G-MDSCs suppressed T cell and NK cell responses by reactive oxygen species-dependent mechanisms [27]. The macrophages and dendritic cells took part in immunological surveillance in ALL [28-30]. Tumor immune surveillance was the result of the combined action of various cells including tumor cells, immune cells, and stroma cells. Distinct molecular subtypes of tumors can be interrelated with a defined immune composition and activation state in the tumor microenvironment. Distinct molecular subtypes of tumors evade immune surveillance in different ways. This is very important for precise immunotherapy that the different molecular mutations lead to different patterns of immune escapes [31]. PAX5 alterations are common in B-ALL [3-5].
The effect of PAX5 mutations on leukemia immunity was not well studied. In our research, by CIBERSORT analysis, we found that there were no statistically difference in the percentages of bone marrow T cells subtypes except follicular helper (Tfh) cells between PAX5 mutations B-ALL and wild type B-ALL. The role of Tfh cells was not explicit, some papers showed that tumor infiltrating Tfh cells were related to good clinical prognosis for breast cancer [32,33]. The proportion of CD8+ T cells, as an important effector cells in the tumor environment, had no significant difference between PAX5 mutations B-ALL and wild type B-ALL. Although PAX5 mutations caused changes of T cell subsets, these changes were insufficient to explain immune tolerance by PAX5 mutations. We turned our attentions to changes of macrophages. Our results showed the changes of macrophages were significant in the tumor environment. The percentages of M0 in B-ALL with PAX5 mutations was higher than that in B-ALL without PAX5 mutations(p<0.001). Macrophages were significant in tumor immune responses. The function of macrophages in tumor immunity was established because chronic inflammation has a bearing on tumor promotion/ progression. Macrophages not only affected the function of CD8+ T-cells and CD4+ T-helper cells [34], but also affected tumor angiogenesis [35].
The function of macrophages depends on the polarization state of macrophages in the tumor microenvironment. M1 macrophages had anti-tumor effects, M2 macrophages can act to directly increase tumor growth. Conversion of M1 to M2 phenotypes suppresses intertumoral CD8+ T cell recruitment [36]. M2 macrophages drove T-cell exhaustion by inhibitory molecules [37]. M0 macrophages are presumed naïve cells that have not been stimulated or received signals that promote activation and functional polarization. M0 and M2 macrophages have radically different characteristics compared with M1 macrophages [38]. The exosomes from M1 macrophages, rather than the exosomes from M0 or M2 macrophages, supported differentiation of mesenchymal stem cells [39]. In recent years, multiple studies showed that high M0 indicated poor prognosis in tumors [40-44]. In the tumor microenvironment, tumor cells induced the immune escape by inhibiting the polarization of M0 to M1 or promoting the polarization of M0 to M2. PAX5 mutations inducing immune escape might be related to PAX5 mutations inhibiting the polarization of M0 macrophages. In addition to changes of immune cells of ALL with PAX5 mutations, we also focused on the genetic changes in the tumor microenvironment of ALL with PAX5 mutations. GO analysis showed GEPs were involved in MHC class II protein complex, MHC class II receptor activity, antigen processing and presentation. GO analysis also showed GEPs were involved in endocytic vesicle membrane, trans-Golgi network membrane. Macrophages are important antigen presenting cells.
The endocytic vesicle and trans-Golgi network played an important role in antigen processing and presentation [23]. The difference in antigen processing and presentation not only affected the functional state of macrophages, but also affected the anti-tumor effect of macrophages [45,46]. GEPs induced by PAX5 mutations might be involved in changes of macrophages antigen presentation and changes of macrophages polarization state. From GO analyses, except MHC class II, PTPs were the main genes involved in BP and MF classifications. PTPs were required for extensive biological processes including antigen presentation [24]. In the tumor microenvironment, PTPs were involved in the intracellular signaling and the intercellular signaling between immune cells, tumor cells and stroma. PTPs participated in antigen receptor signal transduction, macrophages migration and polarizing, T cells activation and angiogenesis [47-50]. Abnormalities in PTPs took part in the pathogenesis of cancer and immune deficiencies [24]. GEPs of PTPs included transmembrane PTPs such as PTPRE, PTPRM, PTPRK, non-transmembrane PTPs such as PTPN1, PTPN2, PTPN12, PTPN23 and Myotubularin MTMR1. The highly expressed genes included PTPRM, PTPN2, PTPN23, MTMR1 and the low expression genes included PTPRE, PTPRK, PTPRE, PTPN1, PTPN12(Supplementary material 2). Current research showed that high expression of PTPRM predicted poor prognosis and promoted tumor growth and lymph node metastasis in cervical cancer and breast cancer [51,52].
The increased PTPN2 expression correlated with tumor progression [53]. The deficiency of PTPN1 is sufficient to promote the development of acute myeloid leukemia [54]. One study suggested that high PTPN2 expression decreased antitumor immune responses in the tumor microenvironment. In several colorectal carcinoma models, the specific PTPN2 deletion in antigen presenting cells or T-cells reduced tumor burden by promoting CD44+ effector/memory T-cells, as well as CD8+ T-cell infiltration and cytotoxicity into the tumor [53]. Another study showed activation of PTPN2 reduced IFN-γ-induced inflammatory responses in macrophages by reducing expression or secretion of multiple cytokines [55]. The cleavage of PTPN2 induced a shift of immune response toward anti-inflammatory state by alteration of cytokine and chemokine profiles [56]. PTPN1 deficiency increased M2 macrophage responses accompanied by increased cytokines IL-10 and IL-4 [54]. PTP1B deficiency increased the effects of proinflammatory in both human and rodent macrophages and induced a loss of viability in resting macrophages [57]. The above-mentioned studies suggested that PTPs were involved in the polarization and migration of macrophages. GEPs of PTPs might participate in changes of macrophages induced by PAX5 mutations. Based on the PPI network and co-expression network, we successfully built 3 highly associated genes, such as PDGFRB, IL1B, CD34, which were marker genes of angiogenesis. Macrophages were supposed to take part in inflammatory and tumor angiogenesis [58].
PDGF signal pathway [59,60] and IL1 signal pathway [61,62] played important roles in tumor angiogenesis caused by macrophages [63]. CD34 was not only an important marker of hematopoietic stem cells, but also an important marker of angiogenesis in tumor microenvironment [64,65]. Current PAX5 studies showed that PAX5 mutations did not reverse B cell differentiation into hematopoietic stem cells and did not affect CD34 expression [66-68]. Marrow angiogenesis was also one of poor prognostic factors of leukemia [69,70]. CD34 staining was still an important method to reflect the micro vessel density (MVD) in leukemia bone marrow microenvironment [69,71]. So, we have reason to infer that high expression of CD34 induced by PAX5 mutations might suggest angiogenesis, which needs to be verified by subsequent experiments. Tumor associated macrophages promoted angiogenesis by production of pro-angiogenic factors [72]. In hepatocellular carcinoma, M0 Macrophage enriched angiogenesis hallmark genes [73]. Combined with other studies and our analysis, we inferred that abnormal macrophage induced by PAX5 mutations were associated with angiogenesis induced by PAX5 mutations by PDGF signal pathway and IL1 signal pathway. In brief, PAX5 mutations led to macrophage changes, and macrophage changes were related to processes of presentation and angiogenesis in leukemia microenvironment.
For more Articles on: https://biomedres01.blogspot.com/



No comments:
Post a Comment
Note: Only a member of this blog may post a comment.